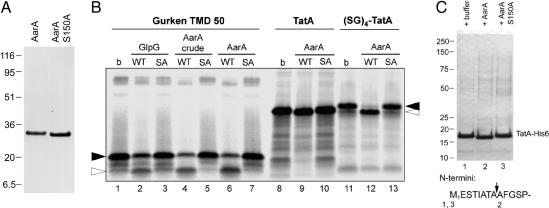
Fig. 3.
In vitro cleavage of TatA by purified AarA protease. (A) Coomassie-stained SDS/PAGE of NiNTA agarose-purified wild-type AarA-His6 (1 μg) and its active-site mutant S150A (1.5 μg). (B) The model substrate derived from Drosophila Gurken (23), full-length Prov. stuartii TatA and its variant, which were N-terminally extended by four SerGly repeats [(SG)4-TatA], were in vitro translated and labeled with [35S]Met. Substrate peptides were incubated with the indicated purified enzymes or crude detergent-solubilized membranes from E. coli expressing AarA (lanes 4 and 5) for 60 min at 30°C, separated by SDS/PAGE, and autoradiographed (buffer only; SA, GlpG-S201A or AarA-S150A). The sizes of the uncleaved substrate and its C-terminal cleavage product are denoted by filled and open arrowheads, respectively. In all cases, the substrate cleavage by AarA depended on the presence of the predicted catalytic serine residue. Cleavage of the Gurken peptide by purified E. coli rhomboid GlpG is shown for comparison. Reactions shown in lanes 2, 3, 6, 7, 9, 10, 12, and 13 contained 3.8 μg of enzyme per 40 μl of reaction, and those in lanes 4 and 5 contained 5.4 μg of enzyme per 40 μl of reaction. (C) Nickel-affinity chromatography-purified TatA-His6 (190 μg/ml) was incubated in the presence of buffer only (b), purified AarA, or its active site mutant S150A (both at 15 μg/ml) at 30°C for 2.5 h. Mixtures were then separated by SDS/PAGE and stained by Coomassie brilliant blue. Sequences of the N termini of the resulting TatA bands were determined by automated Edman degradation. The AarA cleavage site in Prov. stuartii TatA is indicated by an arrow.