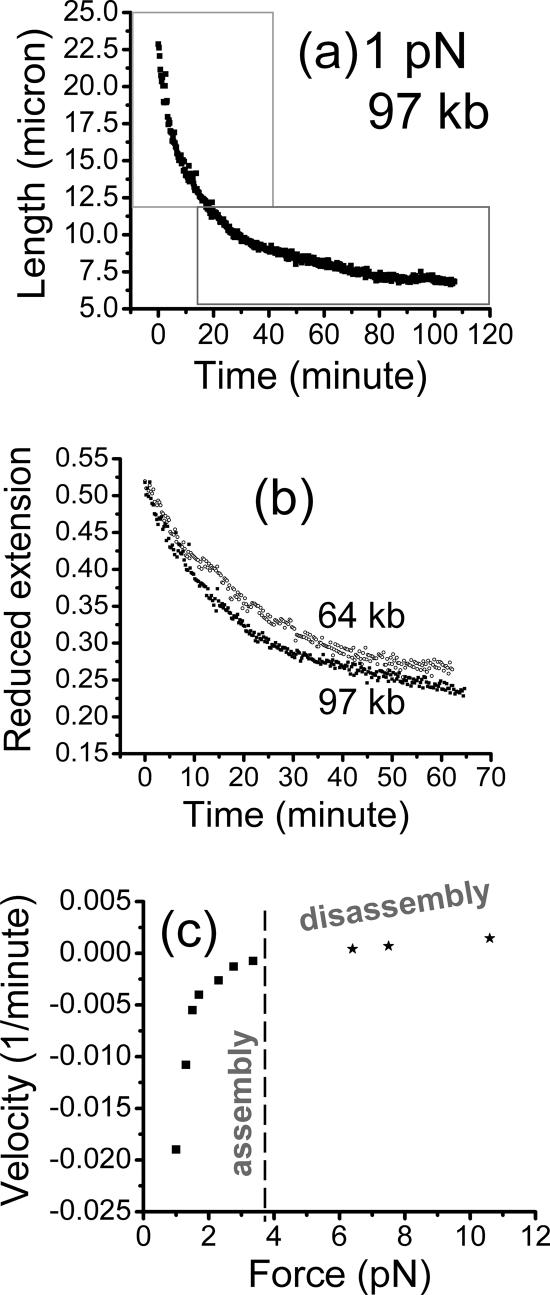
Figure 3.
Assembly of chromatin using diluted Xenopus egg extract, against constant forces, for −ATP (apyrase-treated) conditions. (a) Extension of a 97-kb DNA versus time measured by the vertical MT system, for most of the assembly reaction against a 1-pN force. The initial rapid contraction of the molecule during the early stage of assembly starting at the initial extension of 28 μm was not recorded. Boxes are drawn showing the two regions where exponential fits to the data can be made: the initial time course (top box) fits an exponential with a relaxation time of 10 min; later data in the bottom right box fit a slower relaxation time of 35 min. (b) Extension divided by naked DNA contour length l0 (reduced extension) for different-length assembly runs against forces of 1 pN (filled squares, 97 kb; open circles, 64 kb). Collapse of the curves onto one exponential demonstrates the simple scaling found for the −ATP assembly reaction. (c) Chromatin assembly and disassembly speeds for −ATP conditions show the force dependence expected in a simple force-controlled chemical reaction; figure shows “reduced velocities” (bead velocities divided by naked DNA contour length) of chromatin assembly (negative) and disassembly (positive). A smooth force–velocity relation is obtained, with a zero velocity or stall force of 3.5 pN (dashed line). At this point, assembly and disassembly processes compensate for one another, resulting in no net change in DNA compaction.