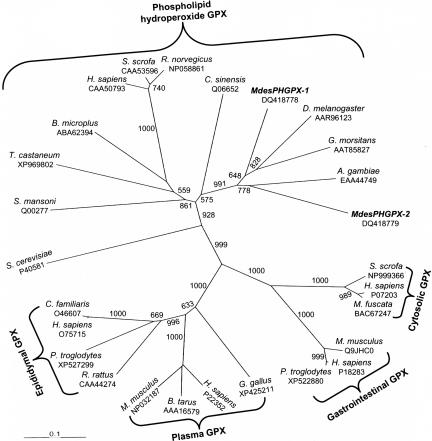
Fig. 1.
Dendrogram of the GPX families calculated from aligned amino acid sequences. The topology and branch lengths of the radial phylogram were produced by the distance/neighbor-joining criteria. Numbers at the branches correspond to bootstrap support >50%. M. destructor phosopholipid hydroperoxide GPXs (PHGPXs) MdesPHGPX-1 and MdesPHGPX-2 group with PHGPXs from other Diptera. Taxa and GenBank accession numbers included are as follows: Saccharomyces cerevisiae, P40581; Schistosoma mansoni, QO0277; Tribolium castaneum, XP_969802; Boophilus microplus, ABA62394; Homo sapiens, CAA50793; Sus scrofa, CAA53596; Rattus norvegicus, NP058861; Citrus sinensis, Q06652; M. destructor, DQ418778; D. melanogaster, AAR96123; G. morsitans, AAT85827; A. gambiae, EAA44749; M. destructor, DQ418779; Sus scrofa, NP999366; H. sapiens, P07203; Macaca fuscata, BAC67247; Mus musculus, Q9JHC0; H. sapiens, P18283; Pan troglodytes, XP_522880; Gallus gallus, XP_425211; H. sapiens, P22352; Bos taurus, AAA16579; Mus musculus, NP032187; Rattus rattus, CAA44274; P. troglodytes, XP_527299; H. sapiens, O75715; Canis familiaris, O46607.