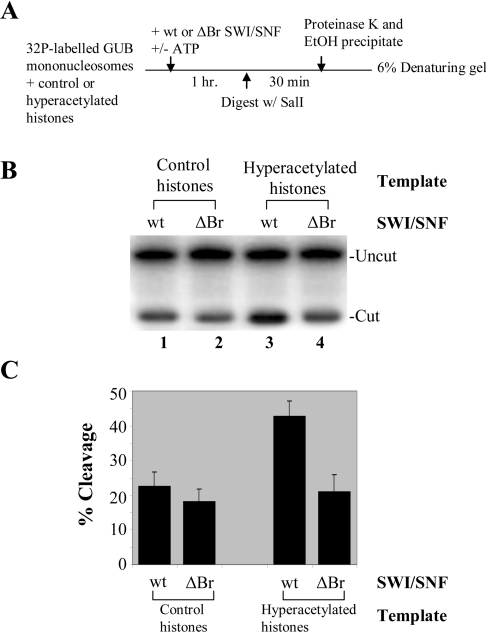
Figure 5. The wild-type SWI/SNF complex has enhanced remodelling activity on hyperacetylated nucleosomes compared with the Δbromodomain SWI/SNF complex.
(A) Diagram of the restriction enzyme accessibility assay as described previously [36,46]. A 32P-labelled GUB fragment was reconstituted into mononucleosomes with either control or hyperacetylated histones for use in this assay. An equal amount of wild-type or Δbromodomain SWI/SNF was added to this GUB template in the presence or absence of ATP followed by SalI digestion. After stopping the digestion reaction and ethanol precipitation, the DNA was resolved on a 6% acrylamide/8 M urea sequencing gel at 150 V for 3 h. The gels were dried and visualized by autoradiography and quantified. (B) Restriction enzyme accessibility assay showing increased activity on hyperacetylated nucleosomes when the Swi2/Snf2 bromodomain is present in the complex. The uncut and cut DNA fragments are labelled. (C) The quantification of three independent experiments is shown as the percentage cleavage of the mononucleosome templates by the restriction enzyme in the presence of the wild-type or the Δbromodomain mutant chromatin-remodelling complex SWI/SNF. The amount of digestion in the absence of ATP is subtracted from that value obtained in its presence when the percentage cleavage was calculated. Quantification of the experiments shows that a similar percentage of the unmodified nucleosome template (with control histones) is cleaved with both the wild-type and the mutant complexes. However, with hyperacetylated nucleosomes as the template, there is a 2-fold increase in remodelling activity of the wild-type SWI/SNF compared with the Δbromodomain SWI/SNF complex.