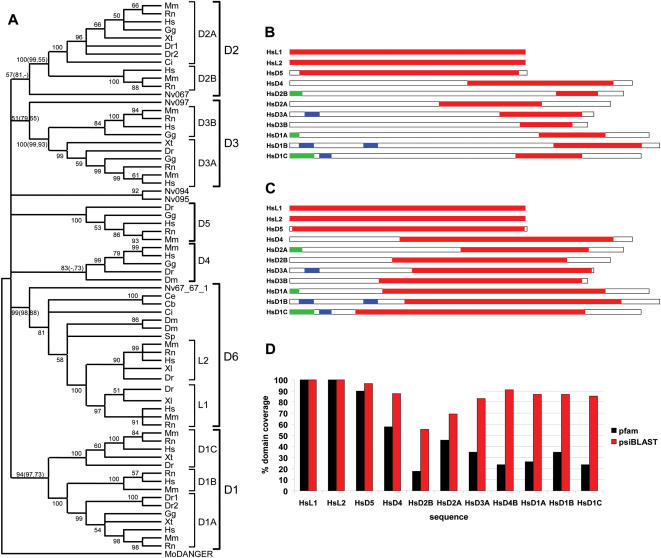
Figure 1. DANGER proteins originated early in metazoan evolution and encode a highly divergent MAB-21 domain.
(A) Neighbor-joining (NJ) consensus tree of the DANGER superfamily defines orthologous relationships among DANGER sequences (D1–D6) from vertebrates (Dr, Danio rerio; Xl, Xenopus laevis; Xt, Xenopus tropicalis; Gg, Gallus gallus; Rn, Rattus norvegicus; Mm, Mus musculus; Hs, Homo sapiens), invertebrates (Ci, Ciona intestinalis; Sp, Strongylocentrotus purpuratus; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; Cb, Caenorhabditis briggsae, Nv, Nematostella vectensis), and the choanoflagellate (Mo, Monosiga ovata). Numbers at branches are bootstrap values from the NJ analysis. Bootstrap and quartet puzzling support values from maximum parsimony (MP) and maximum likelihood (TREE-PUZZLE) analyses, respectively, are given in parentheses. (B) Domain prediction analysis using the Pfam Mab-21 profile, containing four sequences, identifies a partial Mab-21 domain (showed as red boxes) in the human DANGER (HsD1-5) proteins. Predicted signal peptides and transmembrane regions are depicted by green and blue boxes, respectively. (C) Domain prediction analysis, using a Mab-21 profile generated by psi-BLAST containing 63 animal sequences, results in Mab-21 domain extension in all human DANGER proteins. (D) Quantification of Mab-21 domain coverage as predicted by the Pfam and the psiBLAST-generated Mab-21 profiles in all human DANGER proteins. Similar Mab-21 domain extension is observed between orthologous DANGER sequences from other species.