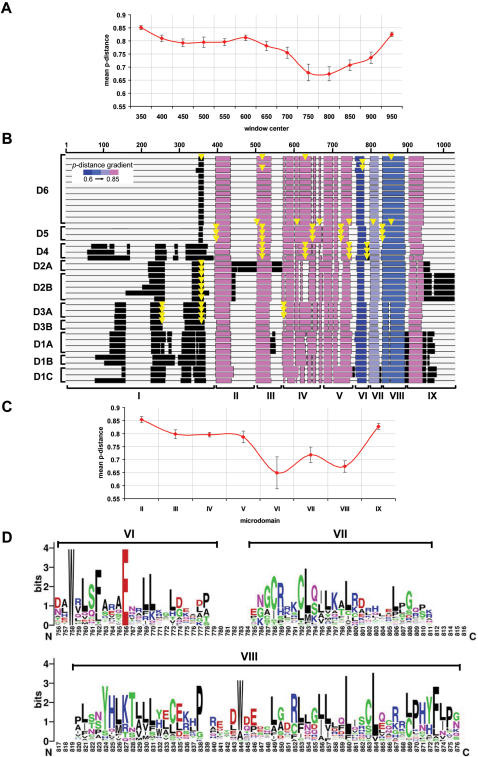
Figure 2. Patterns of sequence evolution along the Mab-21 domain.
(A) Graphical representation of the p-distances calculated along the multiple sequence alignment of representative DANGER proteins with a sliding window of 100 amino acids and a step of 50 amino acids. Bars represent standard errors of mean p-distance values. (B) Multiple sequence alignment of representative DANGER sequences (D1–D6) in block format. Microdomains II to IX are colored according to the mean value of the proportion of amino acid differences (p-distance) among all sequences. Intron positions are mapped as yellow arrows. (B) Graphical representation of the p-distances for each microdomain. Bars represent standard errors of mean p-distance values. (D) Pattern of sequence conservation (logo) along the three most conserved Mab-21 microdomains (VI–VIII).