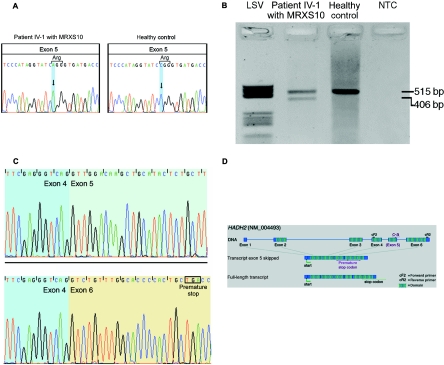
Figure 2. .
A, Silent c.574C→A substitution in exon 5 of HADH2. Left, Sequence of MRXS10-affected patient IV-1, who harbors the c.574C→A substitution. Right, Wild-type sequence of a healthy control. B, RT-PCR amplification of the HADH2 gene through use of primers HADH2-cF2 and HADH2-cR2. In patient IV-1 (lane 2), the amplification results in a 515-bp wild-type fragment and in a second, shorter fragment of 406 bp. In the healthy control (lane 3), only the wild-type 515-bp fragment is visible. The negative template control is shown in lane 4. Molecular weight marker LSV is shown in lane 1. C, cDNA sequences of the exon boundaries (between exons 4 and 5) of the 515-bp fragment with exon 5 (top) and of the exon boundaries (exons 4–6) of the 416-bp fragment lacking exon 5 (bottom). D, Genomic structure of HADH2. The gene consists of six coding exons, which result in a 261-aa protein with a conserved short-chain dehydrogenase domain at amino acid positions 28–257 (hatched boxes). The c.574C→A substitution is located in exon 5. In the transcript variant lacking exon 5, a premature stop codon is created in exon 6. The position of the primers used for cDNA amplification, as shown in panel B, are indicated by “cF2” and “cR2.'