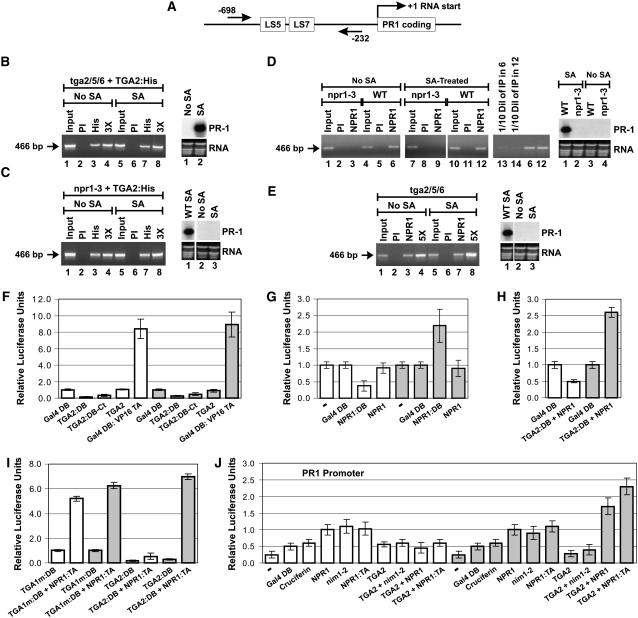
Figure 1.
NPR1 Is a Coactivator Required for Transcriptional Activation by a TGA2-NPR1 Complex in SA-Treated Cells Only.
(A) Graphic representation of the PR-1 gene. The straight arrows and the numbers indicate the positions of the PCR primers used for ChIP experiments. LS5 and LS7 are two DNA regions containing the TGA factor cognate binding sequence TGACG (Lebel et al., 1998).
(B) ChIPs of TGA2:His expressed in tga2/5/6 knockout plants. At right is an RNA gel blot illustrating that the TGA2:His protein complemented the tga2/5/6 mutation and restored PR-1 inducibility.
(C) ChIPs of TGA2:His expressed in npr1-3 mutant (npr1-3) Arabidopsis plants. At right is an RNA gel blot illustrating that the TGA2:His protein expressed in the npr1-3 mutation did not bring about expression of PR-1 regardless of whether tissues were treated or not with SA. An RNA gel blot from wild-type plants treated with SA is shown for comparison.
For (B) and (C), ChIPs were conducted with anti-His antibodies conjugated to agarose beads.
(D) ChIPs of NPR1 from wild-type (NPR1) and npr1-3 mutant (npr1-3) plants. At right is an RNA gel blot illustrating that the PR-1 gene is not expressed in the npr1-3 mutant and that expression in the wild-type plant is dependent upon SA treatment.
(E) ChIPs of NPR1 from tga2/5/6 knockout Arabidopsis plants. At right is an RNA gel blot illustrating that the PR-1 gene is not expressed in the tga2/5/6 mutant regardless of whether tissues have been treated or not with SA. An RNA gel blot from wild-type plants treated with SA is shown for comparison.
For (D) and (E), ChIPs were conducted with anti-NPR1 antibodies for which the specificity has been demonstrated previously (Després et al., 2000).
In (B) to (E), tissues were untreated (No SA) or treated for 6 h with 1 mM SA. PI indicates that ChIP was performed with preimmune serum. PCR was conducted with PR-1 promoter–specific primers. The arrow in each panel indicates the location of the PCR product. The NPR1-3 protein is a deletion version of NPR1 (Cao et al., 1997) that has lost the antigenic region used to raise the anti-NPR1 antibodies used in this study. The inputs represent 2% of the immunoprecipitated material (50-fold dilution). 3X and 5X indicate that the PCR was performed with three and five times the amount of immunoprecipitated material, respectively, to demonstrate that the PCR was in the linear range. In lanes 13 and 14 in (D), one-tenth of the amount of immunoprecipitated material used in lanes 6 and 12, respectively, was used to perform the PCR to demonstrate that the PCR was in the linear range. RNA stained with ethidium bromide is shown for loading comparison.
(F) Histograms illustrating the fact that TGA2 tethered to DNA through Gal4 DB fused to the N terminus (TGA2:DB) or C terminus (TGA2:DB-Ct) does not activate transcription, whereas a chimeric transcription activator composed of the Gal4 DB fused to the transactivation domain of viral protein 16 (Gal4 DB:VP16 TA) does. Gal4 DB represents the baseline level of transcription.
(G) Histograms illustrating the transcription activation of NPR1 tethered to DNA through Gal4 DB (NPR1:DB). NPR1 indicates the absence of fusion. – indicates that only the reporter and internal standard vectors were bombarded into the tissues; no effector was introduced.
(H) Histograms illustrating the effect of NPR1 on the transcriptional activity of TGA2:DB. NPR1 indicates that the protein is expressed without a fusion.
(I) Histograms illustrating the fact that TGA2 tethered to DNA through Gal4 DB (TGA2:DB) interacts very poorly with NPR1:TA in the absence of SA treatment.
In (F) to (I), Arabidopsis leaves were left untreated (white bars) or were treated for 24 h with 1 mM SA (gray bars). The constructs were transfected along with the 5X UASGAL4:firefly luciferase reporter and the CaMV 35S:renilla luciferase internal standard vectors. Data are reported as relative luciferase units. The fold activation represents the relative luciferase units obtained with the given protein or protein pair divided by the relative luciferase units obtained with the unfused Gal4 DB construct alone (baseline transcription). Values are from 25 samples and represent averages ± sd. Every bar represents five bombardments repeated five times (n = 25).
(J) Histograms illustrating the effect of NPR1 and nim1-2 on the transcriptional activity of the TGA2-NPR1 complex. All proteins were native (without fusion), with the exception of NPR1:TA (NPR1 fused to the VP16 transactivation domain), which was used to assess the level of interaction between NPR1 and TGA2 in the context of the PR-1 promoter. The reporter system was the Arabidopsis PR-1 promoter fused to firefly luciferase. The CaMV 35S promoter:renilla luciferase fusion was used as an internal standard. − indicates that no effector was bombarded along with the reporter and internal standard vectors. Cruciferin is an Arabidopsis storage protein used here to illustrate the background level of this system when expressing an unrelated protein. Gal4 DB served the same purpose. Arabidopsis leaves were left untreated (white bars) or were treated for 24 h with 1 mM SA (gray bars). Data are reported as relative luciferase units. Values are from 25 samples and represent averages ± sd. Every bar represents five bombardments repeated five times (n = 25).