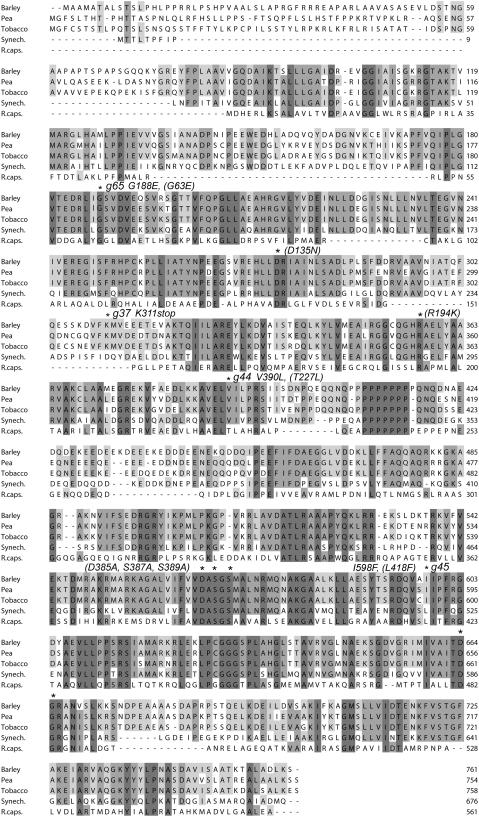
Figure 2.
Alignment of the Amino Acid Sequences of Five Mg-Chelatase D Subunits.
Shadowed boxes highlight invariant and conserved residues. The amino acid residues that were affected by the barley xantha-g37, -g44, -g45, and -g65 mutations are indicated with asterisks as well as other residues that where modified in the study (R. capsulatus residues with numbers are given within parentheses). The MIDAS motif is formed by the invariant amino acid residues Asp-385, Ser-387, Ser-389, Thr-481, and Asp-482 (R. capsulatus numbering). The polypeptides were from barley, pea (Luo and Weinstein, 1997), tobacco (Papenbrock et al., 1997), Synechocystis sp PCC6803 (Jensen et al., 1996), and R. capsulatus (P26175).