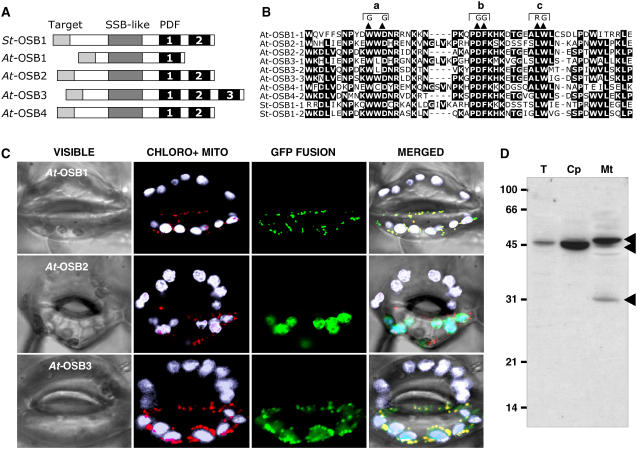
Figure 2.
Characterization of the OSB Protein Family.
(A) Basic structure of OSB proteins from potato (St OSB1) and Arabidopsis (At OSB1 to OSB4). The PDF motifs are numbered.
(B) Alignment of PDF motifs of the proteins. The double substitution mutations (a, b, and c) analyzed in Figure 3 are indicated.
(C) Localization of protein-eGFP fusions in guard cells of N. benthamiana. Green, eGFP fluorescence; white, natural fluorescence of chloroplasts; red, fluorescence of the mitochondrial marker.
(D) Protein gel blot of protein fractions probed with antibodies raised against OSB1: total fraction (T)–, chloroplast (Cp)-, and mitochondria (Mt)- enriched fractions. Protein fractions were extracted from Arabidopsis cells in suspension culture as described (Laloi et al., 2001).