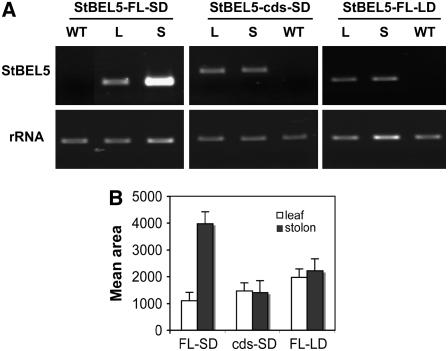
Figure 7.
RNA Accumulation in Transgenic Lines Expressing St BEL5 Transcripts with (St BEL5-FL) and without (St BEL5-cds) the UTRs of the RNA and the Effect of Photoperiod.
Transgenic plants were grown under SDs for 14 d, and the RNA was extracted from 0.5-cm stolon tips (S) and new leaves (L) from three separate plants for each construct (A). For the photoperiod experiment, transgenic plants with the St BEL5-FL construct were grown under LDs for 14 d ([B], St BEL5-FL-LD), and the RNA was extracted from 0.5-cm stolon tips and new leaves from three separate plants. The significant accumulation of St BEL5 RNA in stolon tips shows that signal in the stolon is dependent on photoperiod. One-step RT-PCR was performed using 20 ng of total RNA, a nonplant sequence tag fused to all transgenic RNAs (designated NT-2), and a gene-specific primer from either the open reading frame of St BEL5 (for St BEL-cds) or from the 3′ UTR (for St BEL5-FL). Use of the NT-2 primer makes it possible to discriminate transgenic RNA from native BEL5 RNAs. No PCR product was detected from native St BEL5 RNA extracted from leaves of wild-type plants ([A], WT lanes). The PCR reactions were normalized using rRNA primers. All PCR reactions were standardized and optimized to yield product in the linear range (31 cycles for both BEL5 RNAs and 17 cycles for the rRNA). Expected size of the full-length product was 375 and 550 nucleotides for the cds product. Both constructs were driven by the CaMV 35S promoter in the binary vector pCB201. The 5′ UTR of St BEL5 is 146 nucleotides, and the 3′ UTR is 505 nucleotides. Homogenous PCR products were quantified (B) using ImageJ software (Abramoff et al., 2004) and normalized using the rRNA values. The results of (A) are a representative sample of one of the biological replicates for each treatment. Standard errors of the means of the three biological replicates are shown in (B). Open bars, leaf; closed bars, stolon.