Abstract
The rocG gene of Bacillus subtilis, encoding a catabolic glutamate dehydrogenase, is transcribed by SigL (σ54)-containing RNA polymerase and requires for its expression RocR, a member of the NtrC/NifA family of proteins that bind to enhancer-like elements, called upstream activating sequences (UAS). Unlike the case for other σ54-dependent genes, rocG has no UAS; instead, its expression depends on a sequence located 1.5 kilobases downstream of the rocG promoter, beyond the end of the rocG coding region. The same sequence also serves as the UAS for the downstream rocABC operon and can activate rocG if moved upstream of its promoter. Furthermore, the activating sequence can be moved as far as 15 kilobases downstream of the rocG promoter and still retain partial activity.
Enhancer elements and enhancer-binding proteins are widespread components of transcriptional regulation systems in eukaryotes (1). Enhancer-like elements also have been described in bacteria. In most cases, they are the upstream activating sequences (UAS) required for expression of genes transcribed by RNA polymerases containing σ54 (RpoN)-like subunits (2–5). To initiate transcription, σ54-containing RNA polymerases must interact with additional proteins, members of the NtrC/NifA family, that bind to UAS usually located 100–200 bp upstream of the regulated genes (2–5). It has been shown that UAS can be moved 1–2 kilobases (kb) further upstream from the regulated genes without losing their ability to interact with UAS-binding proteins and to activate expression of the regulated genes (6, 7). A UAS placed downstream of the promoter or even on a separate, catenated, plasmid DNA retains its functional activity in an in vitro assay (8, 9), but no functional bacterial enhancer-like element has yet been found to have a natural location far downstream of the promoter it regulates, despite the fact that this is a common feature of eukaryotic enhancers (1).
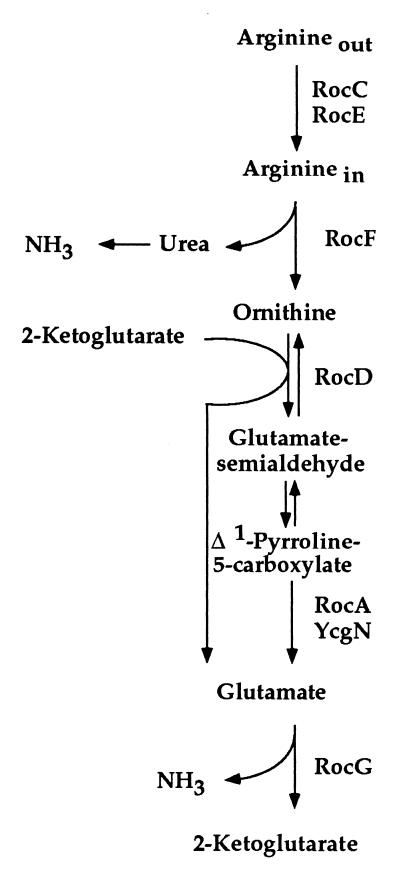
The Bacillus subtilis sigL gene encodes a homolog of the σ54 subunit of RNA polymerase (10), and three B. subtilis UAS-binding proteins, LevR, RocR, and BkdR, have been characterized (11–15). RocR controls two operons, rocABC and rocDEF, involved in arginine degradation to glutamate (Fig. 1). Both operons have SigL-dependent promoters and are induced by arginine, ornithine, or proline (13, 14, 16). In the present work, we show that the B. subtilis rocG gene, encoding a major catabolic glutamate dehydrogenase (GlutDH) (17), is a member of the RocR regulon. RocG activity (glutamate + NAD → 2-ketoglutarate + NH3 + NADH) can be viewed as the final step in the use of arginine, ornithine, and proline as carbon or nitrogen sources (Fig. 1). Two unique features of rocG expression are the location of the apparent RocR binding site downstream of the rocG coding region and the ability of this site to act at distances as far as 15 kb from the rocG promoter.
Figure 1.
The pathway for arginine and ornithine use. The following enzymes are shown on this figure by the names of their corresponding genes: RocC and RocE, putative arginine/ornithine permeases; RocF, arginase; RocD, ornithine aminotransferase; RocA and YcgN, Δ1-pyrroline-5-carboxylate dehydrogenases; RocG, glutamate dehydrogenase. Proline is converted to glutamate via Δ1-pyrroline-5-carboxylate. The proline degradation pathway shares RocA/YcgN- and RocG-dependent steps with the roc-pathway.
MATERIALS AND METHODS
Bacterial Strains and Culture Media.
B. subtilis strains used in this study are derivatives of wild-type strain SMY and were constructed by chromosomal DNA or plasmid transformation as described (17). Cells were grown at 37°C in TSS minimal medium (18) with 0.5% glucose or 0.4% succinate as the carbon source and 0.2% of other carbon or nitrogen sources. Other microbiological procedures were as described (17).
Plasmid and Strain Constructions.
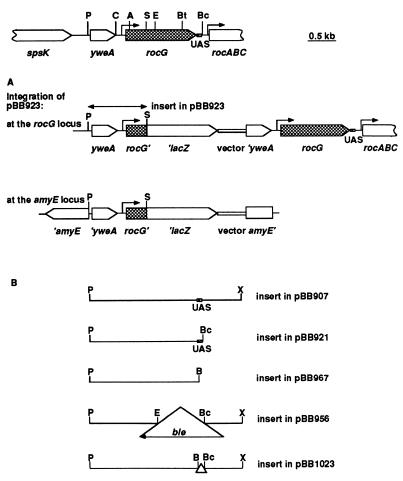
General methods for DNA manipulations and transformation were as described (17). The genetic and restriction maps of the rocG locus and some of the plasmids used in this study are presented in Fig. 2. Plasmids were constructed as described below or by deleting or subcloning fragments of the rocG region (17, 19).
Figure 2.
The genetic map of the rocG region (19) and plasmids carrying different parts of this region. The structure of the chromosome resulting from integration of pBB923 at the rocG or amyE locus is shown. For pBB956, pBB921, and pBB967, such integration events are shown in Fig. 4 (see strains BB1541, 1566, and 1568, respectively). Plasmids pBB907, 956, and 1023 are derivatives of pBB544 (20), and pBB921, pBB923, and pBB967 are derivatives of pJPM82 (20). Plasmid pBB907 was described previously (17). Restriction sites are abbreviated as follows: A, AccI; B, BamHI; Bc, BclI; Bt, BstYI; C, ClaI; E, EcoRI; P, PstI; S, SacII; X, XhoI. The XhoI and BamHI sites were constructed by PCR. The locations of the rocG and rocA promoter regions are indicated by right-angle arrows. The lacZ gene and the vector part of the plasmids are not to the scale.
Construction of rocG-lacZ fusions.
The 1.22-kb PstI-EcoRI fragment (Fig. 2) containing the 5′-part of rocG and 692 bp upstream of the rocG initiation codon was excised from pBB907 (ref. 17; Fig. 2B) with BamHI and EcoRI and was cloned in pJPM82 (20). The resulting plasmid, pBB914, contained the rocG fragment fused to the promoterless Escherichia coli lacZ gene in the proper orientation. pBB923 (Fig. 2A) was created from pBB914 by deleting the 3′-terminal 0.16-kb SacII-EcoRI fragment of the rocG insert. To construct pBB986 and pBB993, the 0.37-kb BstYI-BclI fragment containing the 3′-end of rocG and additional 106 bp of downstream DNA (Fig. 2) was cloned in two orientations in the BamHI site of pBB923 upstream of the PstI-SacII insert. rocG-lacZ fusions were integrated into the chromosome (Fig. 2A) either at the amyE locus by a double-crossover recombination event as described (17) or at the rocG locus by a single-crossover recombination event. The location of fusions at the rocG locus was verified in genetic crosses with linked markers.
Construction of a rocG null mutant.
A deletion–insertion mutation within the rocG gene (pBB956; Fig. 2B) was made in a manner similar to that described for pBB918 (17) by replacing the 0.85-kb EcoRI-BclI fragment of pBB907 (Fig. 2B) with the 1.6-kb EcoRI-BamHI ble cassette, excised from pJPM136 (17). Strain BB1541 containing the Δ(rocG-DAS)∷ble allele was constructed as described for strain BB1284 (ΔrocG∷ble) (17).
Plasmid pBB921 (Fig. 2B) was constructed by cloning the 2.1-kb PstI-BclI fragment cut from pBB907 (Fig. 2B) with BamHI and BclI in the BamHI site of pJPM82 (20) (the direction of rocG transcription in this plasmid coincides with that of the lacZ gene). pBB967 (Fig. 2B) was made in a similar way by cloning a 2.0-kb BamHI-BamHI PCR product that contained the entire rocG gene and 9 bp of the downstream region.
Plasmid pBB1023, lacking the 106 bp of the downstream activating sequence (DAS)/UAS region, was constructed from pBB907 (17) by replacing the 2.1-kb PstI-BclI fragment (as in pBB921) with the 2.0-kb PstI-BamHI fragment of pBB967 (Fig. 2B). Strain BB1664 (ΔDAS/UAS) has the deletion mutation of pBB1023 in the chromosomal rocG rocA locus. It was isolated as a spontaneous derivative of a strain carrying an integrated pBB1023 at the rocG rocA locus in which the plasmid (and its neomycin-resistance gene) had been eliminated by recombination. The replacement of the chromosomal rocG rocA locus by its ΔDAS/UAS allele was confirmed by comparing sizes of the PCR fragments from the wild-type and mutant chromosomal loci.
A rocD∷aphA3∷spc strain was constructed from the rocD∷aphA3 mutant (14) by inactivating the aphA3 marker by a double-crossover recombination event using pBB838. The latter plasmid was constructed by cloning the 1.1-kb blunt-ended spc-cassette, cut from pJL73 (21) with BamHI and EcoRI, into the BlpI site of the aphA3 insert of pDG782 (22). The direction of spc transcription in pBB838 coincides with that of the aphA3 gene.
Enzyme Assays.
β-galactosidase activity was determined as described and was expressed in Miller units (17). GlutDH activity was assayed and expressed in units as described (17).
RESULTS AND DISCUSSION
Identification of the rocG Promoter. The results of genetic analysis placed the rocG promoter between the ClaI site and the rocG initiation codon located upstream of the AccI site (Fig. 2). Integration into the chromosome of a plasmid, carrying an internal AccI-EcoRI fragment of rocG (Fig. 2), disrupted the rocG gene and caused inability to use arginine, ornithine, or proline as sole carbon source, characteristic of a rocG null mutant (17). In contrast, a strain with an integrated plasmid, carrying the ClaI-EcoRI fragment (Fig. 2), retained the wild-type growth phenotype and thus was able to express the rocG gene.
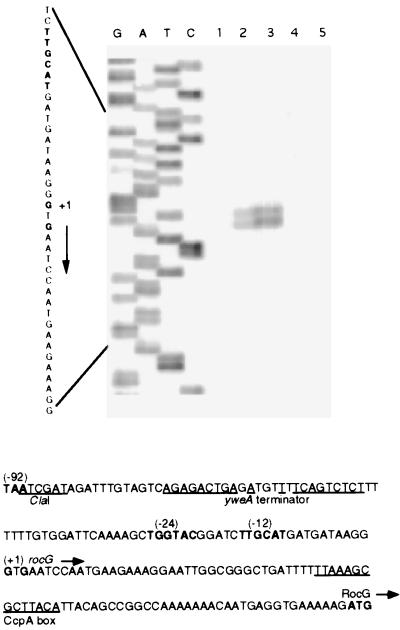
A primer extension experiment identified two closely spaced, apparent transcription start points for rocG at positions G(−87) and G(−85) with respect to the rocG initiation codon (Fig. 3), ≈90 bp downstream of the ClaI site. A putative SigL-dependent promoter had been recognized upstream of the rocG gene (ref. 19; in ref. 19, the rocG gene is referred to as ipa-75d) (Fig. 3). Our primer extension results are consistent with use of this promoter for rocG expression.
Figure 3.
Determination of the rocG transcription start site. Primer oBB56 (5′CTTAATGATTGTTTGGGTAGAC), corresponding to positions +78 to +57 with respect to the rocG initiation codon, was extended (17) with Superscript II reverse transcriptase (GIBCO/BRL) by using total RNA (17) from the following sources: lane 1, B. subtilis strain SMY grown in glucose-ammonia medium; lane 2, strain SMY grown in glucose-ammonia-ornithine medium; lane 3, strain SMY grown with proline as sole carbon and nitrogen source; lane 4, a rocR null mutant grown in glucose-ammonia-ornithine medium; or lane 5, Saccharomyces cerevisiae (Sigma) as templates. The sequence of the nontemplate strand of plasmid pBB916 [contains the PstI-EcoRI fragment (Fig. 2)] obtained by using oBB56 as primer is shown to the left. The sequence of the rocG regulatory region (19) is shown at the bottom. The termination codon of yweA, the −12 and −24 promoter regions, the apparent transcription start points, and a likely initiation codon for the rocG gene are in bold. The directions of transcription and translation are indicated by horizontal arrows. The ClaI site used in plasmid construction, the dyad-symmetry sequence of a putative yweA transcriptional terminator, and a putative CcpA box (cre site) (24, 25) are underlined.
rocG Expression Is Repressed by Glucose, Is Induced by Ornithine and Arginine, and Depends on SigL and RocR. Several rocG-lacZ fusions containing the rocG promoter region but different rocG/lacZ junction points were constructed [pBB923 (Fig. 2A); data not shown] and integrated at the rocG locus of the chromosome, a recombination event that provided the normal chromosomal arrangement of DNA sequences upstream of the rocG part of the fusion. In glucose minimal medium, these fusions were not active (data not shown), but their activity could be detected in the absence of glucose when succinate served as carbon source and ornithine, arginine, or proline was present (Table 1; data not shown). Repression by glucose of the rocG gene, even when ornithine is provided, is mediated by catabolite control protein CcpA (23) (data not shown). A putative CcpA binding site (24, 25) in the rocG regulatory region is indicated in Fig. 3. Expression of the rocG-lacZ fusion in different media correlated with GlutDH activity of RocG (Table 2; data not shown).
Table 1.
Expression of rocG-lacZ fusions
| Strain (genotype) | β-galactosidase activity in different
media
|
|
|---|---|---|
| Succinate Glutamate | Succinate Glutamate Ornithine | |
| rocG-lacZ at the rocG locus | ||
| BB1434 (wild-type) | 1.5 | 10.5 |
| BB1481 (sigL::aphA3) | 2.1 | 1.6 |
| BB1437 (rocRΔ3::aphA3) | 1.2 | 1.4 |
| BB1440 [rocR(T120I)] | 2.8 | 10.2 |
| rocG-lacZ at the amyE locus | ||
| BB1455 (wild-type) | 1.5 | 0.7 |
| BB1458 (rocD::aphA3) | 1.5 | 3.1 |
Strain BB1434 and its derivatives contain a rocG-lacZ fusion [as in pBB923 (Fig. 2A)] at the rocG locus of the chromosome. Strain BB1455 contains the same fusion at the ectopic (unlinked) amyE locus. Null mutations in sigL, rocR, and rocD and the constitutive rocR(T120I) mutation were as described (10, 13, 14). Endogenous β-galactosidase activity in the absence of any rocG fusion was 1-2 units and was not subtracted.
Table 2.
RocG-GlutDH activity
| Strain (genotype) | Carbon and nitrogen sources | RocG activity |
|---|---|---|
| BB1268 (wild-type) | Succinate + glutamate | <40 |
| BB1268 (wild-type) | Succinate + glutamate + ornithine | 649 |
| BB1552 (sigL::aphA3) | Succinate + glutamate + ornithine | <40 |
| BB1553 (rocRΔ3::aphA3) | Succinate + glutamate + ornithine | <40 |
| BB1542 [rocR(T120I)] | Succinate + glutamate | 763 |
| BB1542 [rocR(T120I)] | Succinate + glutamate + ornithine | 829 |
| BB1680 (ahrC::aphA3) | Succinate + glutamate + ornithine | <40 |
A mutation in sigL decreased expression of a rocG-lacZ fusion ≥6-fold, i.e., to near-background levels, in ornithine-containing medium (Table 1), confirming that rocG expression depends in large part on SigL-containing RNA polymerase. No glutamate dehydrogenase activity was detected in a sigL mutant (Table 2).
Expression of rocG-lacZ and accumulation of GlutDH activity also depended on RocR because β-galactosidase and GlutDH activities were reduced ≥7-fold and ≥16-fold, respectively, in a rocR mutant (Tables 1 and 2). Primer extension experiments confirmed that rocG transcription is induced by ornithine or proline and depends on RocR (Fig. 3). In cells containing a constitutive version of RocR, RocR(T120I) (14), the requirement for ornithine as an inducer of rocG expression was at least partially eliminated (Tables 1 and 2). Because rocG expression is induced by arginine, ornithine, and proline and depends on SigL and RocR, rocG belongs to the RocR regulon and is coordinately regulated with the rocA-F genes, involved in degradation of arginine and ornithine to glutamate (Fig. 1). As is the case for other roc genes, expression of rocG depends on the presence of the AhrC protein (Table 2) (14, 26). Because rocG is not induced by glutamate, it is likely that RocG is a dedicated glutamate dehydrogenase responsible for the last step of degradation of arginine and ornithine: i.e., for converting glutamate to 2-ketoglutarate (Fig. 1). An arginine-induced glutamate dehydrogenase also was described in Pseudomonas aeruginosa (27, 28).
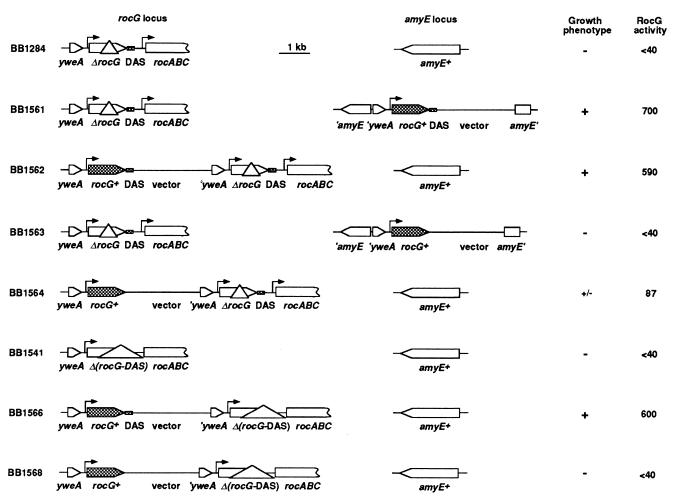
rocG Requires Downstream Sequences for Its Expression. The rocG-lacZ fusions used above were inactive if integrated at the amyE locus of the chromosome: i.e., at a distance of 661 kb from the rocG locus (29) (Table 1; data not shown). A requirement for additional cis-acting regulatory sites, other than the promoter, located upstream of the PstI site or downstream of the SacII site [see insert in pBB923 (Fig. 2A)], could explain inactivity of the rocG-lacZ fusions at the amyE locus. Because we could find no upstream sequence that resembles a putative RocR binding site (13, 16), we placed at the amyE locus a construct containing the entire rocG coding sequence and 106 bp of the downstream sequence [as in pBB921 (Fig. 2B)]. This construct was able to complement the growth defect of the ΔrocG∷ble null mutant and thus contains all elements necessary for rocG expression (Fig. 4, compare strains BB1284 and BB1561). In contrast, a similar construct containing the entire coding region of rocG but only 9 bp of the downstream sequence [as in pBB967 (Fig. 2B)] was not able to complement the rocG mutation (Fig. 4, strain BB1563). We conclude, therefore, that the 106-bp sequence downstream of the rocG gene includes an element required for rocG activation at the amyE locus. We called this element a downstream activating sequence (DAS).
Figure 4.
Requirement of DAS for complementation of rocG mutations. Plasmids pBB921 or pBB967 (Fig. 2B) were integrated in the chromosome of two different rocG deletion mutants (strains BB1284 and BB1541) either at the rocG locus or at the ectopic amyE locus. The full-length rocG allele and the DAS are presented as filled boxes. The vector part of the plasmids (10.5 kb) and the DAS (61 bp) are not to scale. The locations of the rocG and rocA promoter regions are indicated by right-angle arrows. Growth phenotype was scored as ability to use ornithine or proline as sole carbon and nitrogen source. GlutDH activity was determined in succinate-glutamate-ornithine medium.
Both the longer and the shorter constructs [as in pBB921 and pBB967, respectively (Fig. 2B)] were able to complement the growth defect caused by the rocG null mutation when integrated at the rocG locus (Fig. 4, strains BB1562 and BB1564), although, in BB1564, complementation was much less effective. In strain BB1564, the 3′-end of the intact rocG gene is separated from the DAS by ≈13.5 kb. Interestingly, this result implies either that the DAS can act at a distance of ≈15 kb from the rocG promoter (Fig. 4, strain BB1564) or that sequences upstream of rocG, which are absent at the amyE locus but present at the rocG locus, can compensate for the lack of the DAS.
To test whether sequences upstream of rocG can substitute for the DAS, we created a Δ(rocG-DAS)∷ble deletion–insertion chromosomal mutant (Fig. 4, strain BB1541) lacking the 3′-coding region of rocG and 106 bp of the downstream region [as in pBB956 (Fig. 2B)]. As was the case for the previously described rocG∷ble mutation, the new, extended deletion was complemented both at the amyE and rocG loci by a rocG construct containing the DAS (Fig. 4, strain BB1566; data not shown). The Δ(rocG-DAS)∷ble deletion was not complemented by a construct containing the entire coding sequence but lacking the DAS, even when the DAS-less construct was integrated in cis at the rocG locus (Fig. 4, strain BB1568; data not shown). Thus, the sequences upstream of rocG cannot substitute for the lack of the DAS (Fig. 4, strain BB1568). To finally prove the necessity of the DAS for rocG expression, we made a chromosomal deletion of the 106-bp DAS region [as in pBB1023 (Fig. 2B)]. Strain BB1664 (ΔDAS/UAS) containing such a deletion had no detectable GlutDH activity but could be complemented by a DAS-associated rocG gene [as in pBB921 (Fig. 2B)] located at the amyE locus (Table 3).
Table 3.
RocG-GlutDH activity in the absence of the DAS
| Strain (genotype) | RocG activity |
|---|---|
| BB1664 (ΔDAS/UAS) | <40 |
| BB1667 [ΔDAS/UAS amyE::rocG (DAS/UAS+)] | 683 |
| BB1668 [ΔDAS/UAS amyE::rocG (DAS/UAS−)] | <40 |
| BB1665 (ΔDAS/UAS rocD::aphA3::spc) | 65 |
| BB1666 (ΔDAS/UAS rocD::aphA3::spc rocRΔ3::aphA3) | <40 |
Strain BB1664 containing the 106-bp DAS/UAS deletion and its derivatives were grown in succinate-glutamate-ornithine medium. The null mutations in rocD and rocR were described, respectively, in Material and Methods and in ref. 13. All strains carried the ΔgudB::tet mutation to avoid any contribution from GudB-GlutDH (17).
We conclude that high glutamate dehydrogenase activity was detected in strains with integrated rocG plasmids when the DAS was immediately downstream of rocG. Lower, but still significant, activity was seen when the DAS was separated from the rocG promoter by 15 kb of intervening sequence; no activity was detected in the absence of the DAS (Fig. 4). An intermediate GlutDH activity (≈150 units) was detected when the DAS was placed 6.3 kb downstream of the rocG promoter (a plasmid of 4.8 kb containing the 0.45-kb 3′-fragment of rocG without the DAS was integrated into the chromosome of a wild-type strain). In other words, the presence of the DAS in its natural position immediately downstream of rocG (1.5 kb downstream of the promoter) or at a distance as far as 13.5 kb farther downstream of rocG is a prerequisite for rocG expression.
In the rocG-lacZ fusion used in Table 1, the DAS is separated from the rocG promoter by 13 kb of plasmid and rocG sequences (Fig. 2A). This separation presumably explains the low activity of this fusion. The relatively small effect of the rocR(T120I) mutation on rocG-lacZ expression (compare Tables 1 and 2) also could be attributable to separation of the rocG promoter from the DAS.
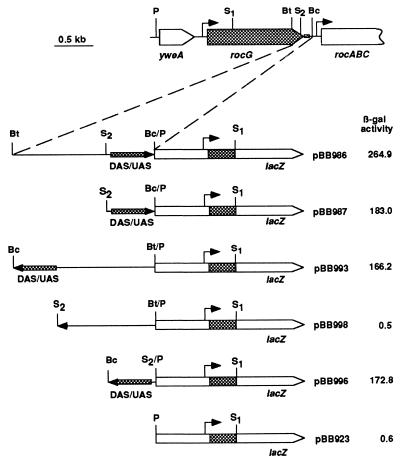
The DAS Overlaps the rocABC UAS and Behaves as an Enhancer. The 106-bp sequence immediately downstream of rocG contains the previously described 61-bp UAS (but not the promoter) of the rocABC operon (Fig. 2). This UAS is a double dyad symmetry element and is a putative binding site for RocR (13, 14, 16). [The phenotypes of strains BB1541 (Fig. 4), BB1664 (Table 3), and their derivatives carrying the rocG (DAS/UAS+) gene at the amyE locus indicate that expression of the rocABC operon, which should be abolished as a result of the UAS deletions, is not required for use of arginine, ornithine, or proline and does not affect GlutDH activity.] To show more directly that this sequence is important for rocG activity and can act as a position- and orientation-independent enhancer element for the rocG gene, we relocated the DAS/UAS upstream of a rocG-lacZ fusion and integrated the construct at the amyE locus. The presence of the DAS/UAS in the upstream position [as in pBB986 (Fig. 5)] greatly stimulated expression of β-galactosidase (compare Tables 1 and 4) [Positioning of the rocABCUAS upstream of another SigL-dependent promoter, levD, also activated its expression (14)].
Figure 5.
The rocG DAS behaves as a position- and orientation-independent enhancer. Derivatives of strain SMY carrying the corresponding rocG-lacZ fusions at the ectopic amyE locus were grown in succinate-glutamate-ornithine medium, and β-galactosidase activity was determined. The lacZ gene and the enlarged fragments of the DAS/UAS region are not to the scale. The arrows show orientation of the DAS/UAS region. The locations of the rocG and rocA promoter regions are indicated by right-angle arrows. The restriction sites are abbreviated as follows: Bc, BclI; Bt, BstYI; P, PstI; S, SacII.
Table 4.
Expression of a rocG-lacZ fusion containing the DAS/UAS in the upstream position
| Strain (genotype) | β-galactosidase activity in different
media
|
|
|---|---|---|
| Succinate Glutamate | Succinate Glutamate Ornithine | |
| BB1501 (wild-type) | 2.1 | 264.9 |
| BB1503 (sigL::aphA3) | 1.2 | 1.2 |
| BB1502 (rocRΔ3::aphA3) | 2.0 | 1.4 |
| BB1522 [rocR(T120I)] | 122.6 | 225.3 |
Strain BB1501 and its derivatives carry a rocG-lacZ fusion [as in pBB986, (Fig. 5)] at the amyE locus.
Expression of the fusion with the DAS/UAS in the upstream position was still subject to glucose repression (data not shown) and depended for its induction on SigL and RocR and arginine, ornithine, or proline (Table 4; data not shown). Note that, with this construct, the constitutive rocR mutation allowed high ornithine-independent expression of the rocG-lacZ fusion, unlike the case in which the DAS/UAS was separated from the rocG promoter by an abnormally great distance (compare Tables 1 and 4). Ability of the DAS/UAS to activate rocG in the upstream position was independent of its orientation or its precise position with respect to the rocG promoter (Fig. 5). These features were previously demonstrated for UAS of other members of the NtrC/NifA family (6, 7, 30–34). The ability of the DAS/UAS to activate rocG from either an upstream or a downstream position makes it very unlikely that stability of rocG mRNA is involved in the activation phenomenon.
The DAS Requirement for rocG Expression Can Be Bypassed.
In otherwise wild-type cells, rocG expression requires the DAS positioned either at its natural location downstream of the gene or relocated upstream or farther downstream of rocG. However, in a rocD mutant, defective in ornithine aminotransferase (Fig. 1), this requirement for the DAS was partially alleviated. In such a mutant, a rocG-lacZ fusion lacking the DAS and integrated at the amyE locus was more active in cells grown in the presence of ornithine (Table 1). Moreover, in the absence of RocD, a full-length rocG gene lacking the DAS [rocD mutant versions of strains BB1563 or BB1664 (Fig. 4 and Table 3)] could provide measurable GlutDH activity (65–120 units). It was shown previously that RocR protein is activated by ornithine and that RocR-dependent expression of the rocDEF operon in ornithine-containing medium is higher in a rocD mutant than in a wild-type strain (14). It seems likely that a defect in ornithine aminotransferase, catalyzing interconversion of ornithine and Δ1-pyrroline-5-carboxylic acid (Fig. 1), leads to intracellular accumulation of ornithine in ornithine-containing medium. Under these conditions, RocR may adopt a more active conformation that allows the protein to bind to low affinity sites or to interact with SigL without binding to DNA. The DAS-independent, ornithine-stimulated expression of rocG still depended on RocR (Table 3). Conditions that allow other proteins of the NtrC/NifA family to activate transcription in the absence of their respective UAS have been extensively described (7, 35–37).
Conclusions and Implications. The presence of a functional enhancer element downstream of a σ54-dependent gene is unique to the B. subtilis rocG gene. The single NtrC binding site upstream of the E. coli glnL gene and immediately downstream of the glnA gene does not contribute to the NtrC-dependent activation of the latter gene in vivo (7), although it can activate glnA if placed upstream of the glnAp2 promoter (38). In most other cases, the corresponding elements are positioned upstream of the regulated promoters (2–5). In several cases of divergently transcribed σ54-dependent genes, enhancer-like elements are found just downstream of the promoters in the 5′ untranslated region or within the N-terminal coding regions of the regulated genes (39, 40). In another case, no UAS could be identified upstream of the argT promoter of Salmonella typhimurium, but neither the presence nor the location of any AS could be ascertained (41). An attempt to move the nifHUAS downstream of the nifH translation start did not result in an active construct (42). The apparent ability of the RocR-DAS complex to influence promoter activity from as far away as 15 kb downstream is an unusual example of enhancer action-at-a-distance.
Another unusual feature of the rocG enhancer element is that it is shared by the rocG gene and the downstream rocABC operon. We imagine that RocR bound to the DAS/UAS interacts independently with molecules of SigL-containing RNA polymerase at the rocG promoter, located ≈1,450 bp upstream, and the rocABC promoter, located ≈100 bp downstream. Both interactions likely require DNA looping (43, 44), although the strengths and geometries of these interactions may be different. In a remarkable case of sequence economy, the DAS/UAS appears also to be the transcription terminator for rocG.
rocG is not completely coregulated with the other roc genes. In contrast to the rocG gene, which is repressed in glucose-containing media by CcpA, the rocABC and rocDEF operons are highly expressed in minimal glucose medium (13, 16) and are not significantly regulated by CcpA (data not shown). This differential regulation of the genes of the roc regulon could reflect different functional roles of the corresponding products. Although RocF and RocD liberate nitrogen groups from arginine and ornithine, respectively, RocG has a dual role, providing both ammonia as a nitrogen source and 2-ketoglutarate as a carbon source (Fig. 1), and may therefore be subject to more complex regulation. Moreover, full RocG activation could lead to depletion of the pool of glutamate, a major cellular anion (45); cells may only allow glutamate degradation in the absence of rapidly metabolizable carbon sources. It will be interesting to find out whether the downstream position of the rocG DAS is a prerequisite for differential regulation of the rocG and other roc genes.
Acknowledgments
We are grateful to C. Alén, C. Jourlin, M. Ratnayake-Lecamwasam, N. Mani, and K. Matsuno for helpful discussions, to M. Débarbouillé, R. Gardan, and G. Rapoport for discussions and gifts of strains, and to M. Débarbouillé, S. Kustu, L. Reitzer, and A. Wright for critical reading of the manuscript. This work was supported by U.S. Public Health Service research grant GM36718.
ABBREVIATIONS
- UAS
upstream activating sequence
- GlutDH
glutamate dehydrogenase
- DAS
downstream activating sequence
- kb
kilobase
References
- 1.Müller M M, Gerster T, Schaffner W. Eur J Biochem. 1988;176:485–495. doi: 10.1111/j.1432-1033.1988.tb14306.x. [DOI] [PubMed] [Google Scholar]
- 2.Kustu S, Santero E, Keener J, Popham D, Weiss D. Microbiol Rev. 1989;53:367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kustu S, North A K, Weiss D. Trends Biochem Sci. 1991;16:397–402. doi: 10.1016/0968-0004(91)90163-p. [DOI] [PubMed] [Google Scholar]
- 4.Morett E, Segovia L. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merrick M J. Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 6.Buck M, Miller S, Drummond M, Dixon R. Nature (London) 1986;320:374–378. [Google Scholar]
- 7.Reitzer L J, Magasanik B. Cell. 1986;45:785–792. doi: 10.1016/0092-8674(86)90553-2. [DOI] [PubMed] [Google Scholar]
- 8.Ninfa A J, Reitzer L J, Magasanik B. Cell. 1987;50:1039–1046. doi: 10.1016/0092-8674(87)90170-x. [DOI] [PubMed] [Google Scholar]
- 9.Wedel A, Weiss D S, Popham D, Dröge P, Kustu S. Science. 1990;248:486–490. doi: 10.1126/science.1970441. [DOI] [PubMed] [Google Scholar]
- 10.Débarbouillé M, Martin-Verstraete I, Kunst F, Rapoport G. Proc Natl Acad Sci USA. 1991;88:9092–9096. doi: 10.1073/pnas.88.20.9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Débarbouillé M, Martin-Verstraete I, Klier A, Rapoport G. Proc Nat Acad Sci USA. 1991;88:2212–2216. doi: 10.1073/pnas.88.6.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin-Verstraete I M, Débarbouillé M, Klier A, Rapoport G. J Mol Biol. 1994;241:178–192. doi: 10.1006/jmbi.1994.1487. [DOI] [PubMed] [Google Scholar]
- 13.Calogero S, Gardan R, Glaser P, Schweizer J, Rapoport G, Débarbouillé M. J Bacteriol. 1994;176:1234–1241. doi: 10.1128/jb.176.5.1234-1241.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardan R, Rapoport G, Débarbouillé M. Mol Microbiol. 1997;24:825–837. doi: 10.1046/j.1365-2958.1997.3881754.x. [DOI] [PubMed] [Google Scholar]
- 15.Débarbouillé M, Gardan R, Arnaud M, Rapoport G. J Bacteriol. 1999;181:2059–2066. doi: 10.1128/jb.181.7.2059-2066.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardan R, Rapoport G, Débarbouillé M. J Mol Biol. 1995;249:843–856. doi: 10.1006/jmbi.1995.0342. [DOI] [PubMed] [Google Scholar]
- 17.Belitsky B R, Sonenshein A L. J Bacteriol. 1998;180:6298–6305. doi: 10.1128/jb.180.23.6298-6305.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fouet A, Sonenshein A L. J Bacteriol. 1990;172:835–844. doi: 10.1128/jb.172.2.835-844.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glaser P, Kunst F, Arnaud M, Coudart M-P, Gonzales W, Hullo M-F, Ionescu M, Lubochinsky B, Marcelino L, Moszer I, et al. Mol Microbiol. 1993;10:371–384. [PubMed] [Google Scholar]
- 20.Belitsky B R, Gustafsson M C U, Sonenshein A L, von Wachenfeldt C. J Bacteriol. 1997;179:5448–5457. doi: 10.1128/jb.179.17.5448-5457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeDeaux J R, Grossman A D. J Bacteriol. 1995;177:166–175. doi: 10.1128/jb.177.1.166-175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guérout-Fleury A-M, Shazand K, Frandsen N, Stragier P. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 23.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 24.Weickert M J, Chambliss G H. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hueck C, Hillen W. Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 26.Miller C M, Baumberg S, Stockley P G. Mol Microbiol. 1997;26:37–48. doi: 10.1046/j.1365-2958.1997.5441907.x. [DOI] [PubMed] [Google Scholar]
- 27.Früh R, Haas D, Leisinger T. Arch Microbiol. 1985;141:170–176. doi: 10.1007/BF00423280. [DOI] [PubMed] [Google Scholar]
- 28.Park S-M, Lu C-D, Abdelal A T. J Bacteriol. 1997;179:5300–5308. doi: 10.1128/jb.179.17.5300-5308.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, et al. Nature (London) 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 30.Birkmann A, Böck A. Mol Microbiol. 1989;3:187–195. doi: 10.1111/j.1365-2958.1989.tb01807.x. [DOI] [PubMed] [Google Scholar]
- 31.Inouye S, Gomada M, Sangodkar U M X, Nakazawa A, Nakazawa T. J Mol Biol. 1990;216:251–260. doi: 10.1016/S0022-2836(05)80317-1. [DOI] [PubMed] [Google Scholar]
- 32.Claverie-Martin F, Magasanik B. Proc Natl Acad Sci USA. 1991;88:1631–1635. doi: 10.1073/pnas.88.5.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin-Verstraete I M, Débarbouillé M, Klier A, Rapoport G. J Mol Biol. 1992;226:85–99. doi: 10.1016/0022-2836(92)90126-5. [DOI] [PubMed] [Google Scholar]
- 34.Weiner L, Brissette J L, Ramani N, Model P. Nucleic Acids Res. 1995;23:2030–2036. doi: 10.1093/nar/23.11.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider B L, Shiau S-P, Reitzer L J. J Bacteriol. 1991;173:6355–6363. doi: 10.1128/jb.173.20.6355-6363.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brahms G, Brahms S, Magasanik B. J Mol Biol. 1995;246:35–42. doi: 10.1016/s0022-2836(95)80067-0. [DOI] [PubMed] [Google Scholar]
- 37.North A K, Kustu S. J Mol Biol. 1997;267:17–36. doi: 10.1006/jmbi.1996.0838. [DOI] [PubMed] [Google Scholar]
- 38.Reitzer L J, Movsas B, Magasanik B. J Bacteriol. 1989;171:5512–5522. doi: 10.1128/jb.171.10.5512-5522.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mullin D A, Newton A. J Bacteriol. 1989;171:3218–3227. doi: 10.1128/jb.171.6.3218-3227.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minchin S D, Austin S, Dixon R A. Mol Microbiol. 1988;2:433–442. doi: 10.1111/j.1365-2958.1988.tb00049.x. [DOI] [PubMed] [Google Scholar]
- 41.Schmitz G, Nikaido K, Ames G F-L. Mol Gen Genet. 1988;210:108–117. doi: 10.1007/BF00331311. [DOI] [PubMed] [Google Scholar]
- 42.Buck M, Woodcock J, Cannon W, Mitchenall L, Drummond M. Mol Gen Genet. 1987;210:140–144. doi: 10.1007/BF00337770. [DOI] [PubMed] [Google Scholar]
- 43.Buck M, Cannon W, Woodcock J. Mol Microbiol. 1987;1:243–249. doi: 10.1111/j.1365-2958.1987.tb00518.x. [DOI] [PubMed] [Google Scholar]
- 44.Su W, Porter S, Kustu S, Echols H. Proc Natl Acad Sci USA. 1990;87:5504–5508. doi: 10.1073/pnas.87.14.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whatmore A M, Chudek J A, Reed R H. J Gen Microbiol. 1990;136:2527–2535. doi: 10.1099/00221287-136-12-2527. [DOI] [PubMed] [Google Scholar]