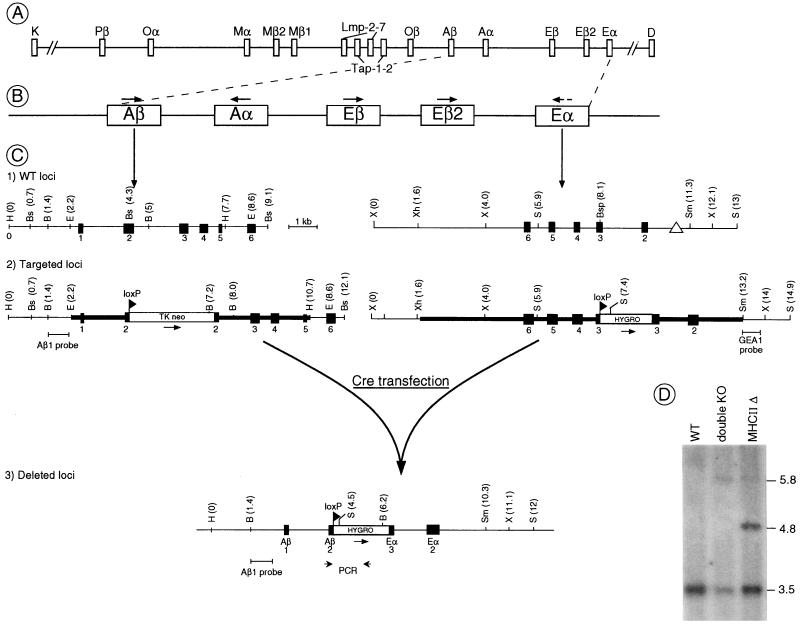
Figure 1.
Strategy for genomic deletion of all classical MHC-II genes. (A and B) Genomic organization of the mouse MHC around and in the class II region. Arrows indicate the transcriptional orientation of the different genes. (C) Mutation strategy. (C1) The wild-type Aβ and Eα genes. Exons are shown as filled boxes. The triangle represents the promoter deletion present in the Eα gene of the H2b haplotype. H, HindIII; Bs, BstEII; Bsp, BspEI; E, EcoRI; B, BglII; X, XbaI; S, SpeI; Sm, SmaI; Xh, XhoI. (C2) The mutated Aβ and Eα loci after homologous recombination. The homologous recombination event introduced a TK-neo cassette into exon 2 of the Aβ gene and a loxP-hygromycin cassette into exon 3 of the Eα gene. The targeting DNA fragments used for electroporation are indicated by a bold line, the loxP sites as triangles. The probes used for the screening are shown under the targeted loci. (C3) Map of the deleted locus after Cre-mediated recombination. The Southern blot probe and PCR primers used to identify the deleted locus are shown under the map. (D) Southern blot analysis of genomic DNA isolated from wild-type embryonic stem (ES) cells and the double-targeted ES clone before and after the deletion event (BglII digest; Aβ1 probe).