Figure 2.
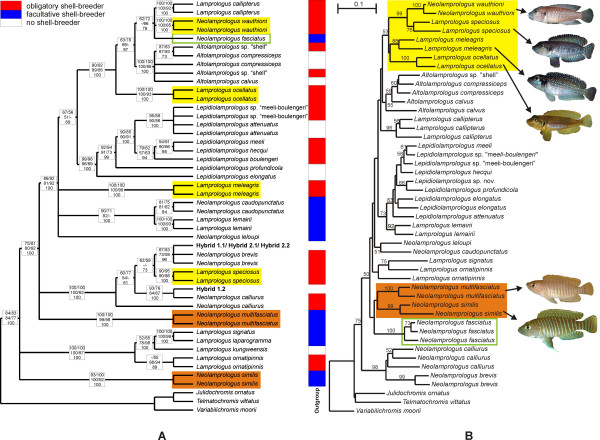
Phylogenetic relationships among the "ossified group" lamprologines. The incongruency between current genus assignments and phylogenetic relationships and the consequent need of a taxonomic revision has been addressed previously [5, 6, 8]. Here, the nomenclature of species in the genus Lepidiolamprologus follows [8] (A) Strict consensus tree of the results obtained from maximum parsimony analysis (MP; 48 most parsimonious trees; tree length, 1390 evolutionary steps; CI excluding uninformative characters, 0.6014; RI, 0.7503; RC, 0.4513), neighbour-joining (NJ), maximum likelihood (ML) and Bayesian inference (BI) based on ND2 sequence data of 48 taxa, representing 27 species of the "ossified group" of lamprologines, four putative hybrid specimens and three outgroup taxa. Boxes on the branches contain bootstrap values obtained from NJ and MP (upper line), ML-bootstrap and quartet puzzling values (middle line), and Bayesian posterior probabilities (bottom line). Only values higher than 50 are shown. Colored bars code for different breeding behaviors. (B) NJ tree based on Nei and Li's distances of the AFLP data of 47 taxa, representing 26 species of the "ossified group" of lamprologines and three outgroup taxa. Bootstrap values > 50 are shown above the branches. The photographs illustrate the phenotypic similarity between Lamprologus meleagris, L. ocellatus, L. speciosus and Neolamprologus wauthioni, and between N. similis and N. multifasciatus. Colored boxes mark eco-morphologically similar taxa with incongruent positions in the mitochondrial and nuclear phylogenies, as well as the disparate placement of N. fasciatus.
