Fig. 3.
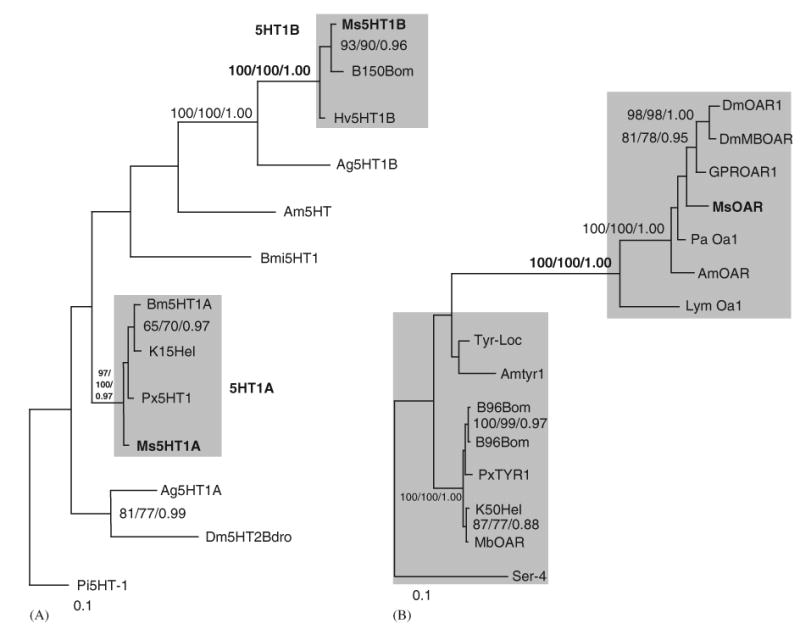
(A) Phylogenetic tree comparing the sequences for Ms5HT1A and Ms5HT1B with receptor sequences for invertebrate 5HT1 receptors. The best Bayesian topology is shown. The shaded boxes denote the two 5HT1 lepidopteran clades. The Ms5HT1A and B sequences are bolded as are relevant values for nodes supporting the two clades. This tree is displayed rooted by P. interuptus. Protein sequences used; Ag5HT1A (EAA04158), Ag5HT1B (EAA13071), genbank:Am5HT (XP_393915), Bmi5HT (AY323235.1), genbank:B150Bom (CAA64862), Dm5HTdro2B (Z11490.1), K15Hel (CAA64863), Pi5-HT1 (AY528822.1), Px5HT1 (BAD72868). Scale bar indicates 10 changes per 100 alignment positions. (B) Phylogenetic tree comparing the amino acid sequence of MsOAR with OA and tyramine receptors from other insects. Shaded areas indicate a cluster of tyramine receptor-like proteins (lower) and a cluster of OA receptor-like proteins (upper). The best Bayesian tree topology is shown. Support values for nodes are shown in the order of maximum likelihood (ML) bootstrap values, ML-corrected neighbor joining bootstrap values and Bayesian posterior probabilities. Only nodes supported by better than 0.85 posterior probability or 65% bootstrap support in 2/3 methods are shown. The M. sexta OAR sequence is bolded, as are the support values for the node separating the OA and the tyramine receptor clades. This tree should be treated as unrooted and is rooted arbitrarily on the C. elegans Tyr sequence for display purposes only. Protein sequences used; GPROAR1 (EAA06361), GPRTYR (EAA07468), AmOAR (NP_001011565), Amtyr1 (CAB76374), B96Bom (CAA64865), Ser-4 (NP_497452), DmOAR1 (CAB38026), DmMBOAR (AAC17442), K50Hel (CAA64864), Lym OA1 (AAC61296), Tyr-Loc (Q25321), MbOAR/Tyr (AAK14402), Pa OA1 (AAP93817), PxTYR1 (BAD72869).
