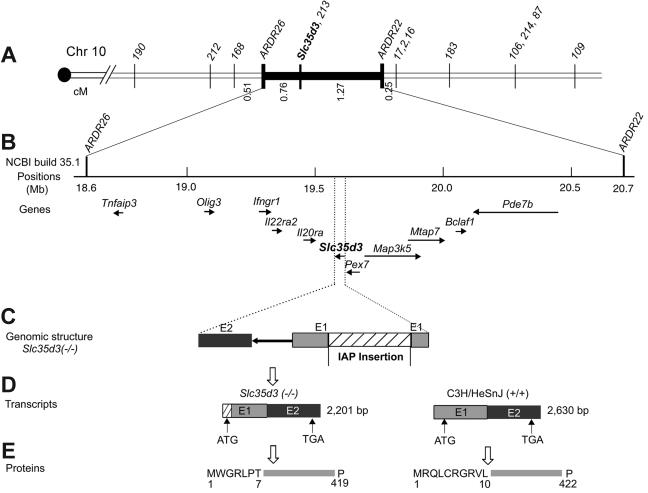
Figure 3.
Identification of the Slc35d3 gene by positional cloning. (A) High-resolution genetic map of the region surrounding the Slc35d3 gene on mouse chromosome 10. Numbered markers are polymorphic MIT microsatellites. ARDR26 (amplified with primers 5′-GGGTGGAGCGGTACTATTCA-3′ and 5′-TGTCTTCAGAGACAACAATCCAG-3′) and ARDR22 (amplified with primers 5′-TTTTCTTTAGGTGGTTTTCTACAGC-3′ and 5′-AGGACAGACAGAGCCCTGAG-3′) are polymorphic microsatellites identified by genome scanning. (B) High-resolution physical map [based on the National Center for Biotechnology Information map viewer (build 35.1)]. The positions of 11 known genes in the critical genetic interval are indicated with their transcription orientations indicated by arrows. (C) The region of the genome from the beginning of exon 1 to the end of exon 2 of Slc35d3. A 5.5-kb IAP element is inserted between nucleotides 12296009 and 12296010 (Contig NT_039 492.5 of Mus musculus build 35.1) in exon 1 of Slc35d3 in the ash-Roswell mutant (Slc35d3−/−). (D) Transcripts of Slc35d3 in ash-Roswell (Slc35d3−/−) and control C3H/HeSnJ brains. Slc35d3 m-RNA of ash-Roswell mutants contains a new and in-frame ATG start site derived from the IAP element. More detailed views of normal and mutant Slc35d3 transcripts are given in Figure 4. (E) Predicted N-terminal sequences of mutant and control Slc35d3 proteins. The substitution of 7 new N-terminal mutant amino acids for the original 10 N-terminal wild-type amino acids is indicated.