Abstract
Crosses between two recently diverged rodent species of the genus Peromyscus result in dramatic parent-of-origin effects on growth and development. P. maniculatus females crossed with P. polionotus males yield growth-retarded conceptuses, whereas the reciprocal cross results in overgrowth and lethality. These hybrid effects are particularly pronounced in the placenta. We previously detected linkage to two regions of the genome involved in the overgrowth effects. One locus, termed Peal, is a paternally expressed autosomal locus mapping to a domain whose house mouse equivalent contains several clusters of imprinted genes. The other locus, termed Mexl, maps to a gene-poor region of the X chromosome. Here we use an advanced intercross line to verify and narrow the regions of linkage and identify candidate genes for Mexl and Peal. While we have previously shown that Mexl affects both pre-and postnatal growth, we show here that Peal affects only prenatal growth. Utilizing criteria such as mutant phenotypes and allelic expression, we identify the loci encoding the homeobox protein Esx1 and the zinc-finger protein Pw1/Peg3 as candidates. Both loci exhibit expression changes in the hybrids.
Introduction
Rodents of the genus Peromyscus are the most common indigenous North American mammals. Peromyscus maniculatus, the deer mouse, is the most widely distributed of the group, ranging from Alaska to Mexico City, from sea level to 14,000 feet elevation, and from the Pacific to Atlantic Oceans (Dewey and Dawson 2001). Despite near ubiquity in North America, P. maniculatus is largely excluded from the southeast United States. This region is inhabited by a recently diverged sister species, P. polionotus (“oldfield mouse”).
Crosses between captive strains of P. maniculatus (BW) and P. polionotus (PO) yield striking parent-of-origin effects on growth (Dawson 1965; Rogers and Dawson 1970). Female BW animals crossed with PO males (bw × po) produce animals that exhibit prenatal growth retardation relative to the parental strains. The bw × po growth effects are particularly pronounced in the placenta, which average half the size of those of either parental genotype. The undergrowth persists through the life of the bw × po offspring, which are fertile and healthy.
Conversely, offspring of female PO animals crossed with BW males (PO × BW) exhibit placental and somatic overgrowth. A variable suite of phenotypes accompanies the PO × BW overgrowth; these range from conceptuses where there is no embryo proper (i.e., only placental and extra-embryonic tissues are present) to those that survive to late gestation. Typically the latter are deformed and oversized to the point that the mother dies at parturition. The very few PO × BW that have survived postnatally have been oversized and female (Dawson et al. 1993). These Peromyscus crosses represent a rare case in which the genetics of mammalian reproductive isolation may be examined.
The parent-of-origin effects seen in the F1 crosses suggested that genes whose expression is dependent on parental origin might underlie the hybrid dysgenesis. Such loci include mitochondrial, sex chromosome-linked, and genes subject to genomic imprinting. Mitochondrial-nuclear genotype interactions have been ruled out as a source of the effects (Dawson et al. 1993). Imprinted genes are autosomal genes that show preferential expression of one parental allele over the other. Many imprinted genes are involved in growth pathways and are expressed in the placenta (Reik et al. 2003).
The X chromosome also shows parental gene expression bias. For example, the paternal copy is preferentially silenced in house mice (Mus musculus) and bovine extra-embryonic tissues (Takagi and Sasaki 1975; Xue et al. 2002). Moreover, the X chromosome is known to contain a disproportionately large number of placental genes (Khil et al. 2004; Moore et al. 2005).
To investigate the genetics of the overgrowth, we initially performed two backcrosses. In the first backcross we mated F1 females (bw × po) with BW males (F1 × BW hereafter). The second cross series consisted of female PO animals crossed with F1 males (PO × F1). We expected both backcrosses to mimic the overgrowth phenotypes seen in the PO × BW F1cross, because both could contribute PO alleles maternally and BW alleles paternally.
We mapped two loci associated with overgrowth through genotypic analysis of the offspring from the two crosses: one linked to the paternally expressed gene Pw1/Peg3, and one linked to the X chromosome (Vrana et al. 2000). We showed that X chromosome loci are imprinted (maternally expressed) in Peromyscus extra-embryonic tissues as in Mus. Thus, both regions of linkage were consistent with the parent-of-origin effects.
We have termed the X-linked locus Mexl (Maternal Expressed X-linked Locus) and the Pw1/Peg3-linked locus Peal (Paternal Expressed Autosomal Locus). Both backcrosses could contribute the PO allele of Mexl maternally and the BW allele of Peal paternally to produce overgrowth. The linkage to each locus, however, is revealed in only one of the two backcrosses, depending on whether the paternal or maternal genome is varied in that cross.
We also previously performed a cross between F1 (bw × po) females × F1 (bw × po) males (Vrana et al. 2000). As expected, this cross yielded small, normal, and oversized offspring. Furthermore, the F1 × F1 offspring genotypes suggested a negative interaction between the two mapped loci (Mexl and Peal). That is, when both parental genotypes varied, we saw a significant reduction in those that had a maternally derived PO Mexl allele and those that had a paternally derived BW Peal allele.
While both backcrosses (PO × F1 and F1 × BW) did in fact yield both normal and oversized offspring, the PO × F1 cross better recapitulated the F1 phenotypes. The offspring of the PO × BW cross exhibited extreme placental and somatic overgrowth, multiple developmental abnormalities, and loss of imprinting (LOI) of certain genes (Vrana et al. 1998). LOI is characterized by a switch from monoallelic to biallelic expression, indicating a perturbation in control of genomic imprinting. In contrast, the offspring of the F1 × BW cross exhibited mild placental/somatic overgrowth, little embryonic lethality or morphologic abnormalities, and no discernible LOI.
We examined the disparity between these two crosses and have provided genetic evidence for a maternal effect involved in the LOI (Duselis et al. 2005). This maternal effect appears to require PO homozygosity in mothers of affected offspring and to be due to a small number of loci. The maternal effect appears to be a separate mechanism contributing to the hybrid overgrowth. Thus, we hypothesize that the combination of the maternal effect and the Mexl-Peal interaction are responsible for the full hybrid overgrowth and suite of associated characteristics.
To further explore the genetics of the hybrid overgrowth, we have confirmed and refined the areas of linkage through a combination of backcrosses and intercrosses. Because no Peromyscus genome is yet mapped, we developed additional genetic markers for these regions. We surveyed these areas for candidate loci and investigated several through sequence identity and gene expression. We focused particularly on Pw1/Peg3 and Esx1 because they were the first candidate genes identified for Peal and Mexl (respectively) and because they remained candidates through the genetic analyses.
Materials and methods
Animal breeding/husbandry
We purchased BW and PO animals from the Peromyscus Genetic Stock Center (http://www.stkctr.biol.sc.edu/). We bred BW females with PO males as the basis of the advanced intercross line. These hybrid animals were then interbred for 2 more generations in the case of the G3 animals, 9 more generations in the case of the G10s, and 12 more generations in the case of G13s. Animals picked to breed for the next generation were chosen randomly. Animals were housed with food and water ad libitum on a 16:8 light:dark cycle.
We collected embryos and placentas from pregnant females, using morphologic criteria and (when applicable) time since birth of previous litters to ascertain developmental stage. Only placentas from pregnancies determined to be past 14.5 days gestation (i.e., mature placental weight) were used for comparisons with results from other crosses. The upper weight limit for placental weights in the parental strains was 0.25 g (Table 1). To allow for any unseen parental variation, we considered any placenta ≥0.3 g overgrown.
Table 1.
Placental weights (in grams) of Peromyscus crosses
| Cross | Range | Mean | SD |
|---|---|---|---|
| BW × BW | 0.078–0.241 | 0.181 | 0.043 |
| PO × PO | 0.098–0.250 | 0.181 | 0.036 |
| BW × PO | 0.038–0.145 | 0.096 | 0.027 |
| PO × BW | 0.232–0.776 | 0.511 | 0.149 |
| G10/13 × BW | 0.127–0.673 | 0.277 | 0.091 |
| PO × G3/8 | 0.102–0.612 | 0.257 | 0.114 |
Backcross data include all mature placentas from this study. Parental and F1 data from Duselis et al. (2005).
Development of markers and genotyping
Genes used as markers were chosen based on their location in the linkage areas. Mus and Homo sequences were obtained from ENSMBL (http://www.ensembl.org) and aligned. Areas in the 5′ or 3′ untranslated regions (UTRs) or small exons and introns that had a high level of conservation were used to design primers. Sequence was obtained from the PO and BW strains by PCR and cloned into the pCR 2.1-TOPO vector (Invitrogen Life Technologies, Carlsbad, CA). Several clones of each allele were sequenced to obtain the most accurate sequence. Sequences were then examined for restriction fragment length polymorphisms (RFLPs) using Vector NTI or TACG (http://workbench.sdsc.edu).
We isolated genomic DNA from embryonic tissues using the Qiagen whole-tissue kit (Qiagen, Valencia, CA) and genotyped them by PCR RFLP analysis. In addition, some Prdx4 genotypes were ascertained by direct sequencing of PCR products amplified with the primers listed below. Digested PCR products were electrophoresed on 7.5% acrylamide gels and scored as BW, PO, or heterozygotes. Pw1/Peg3, Myod1, and Xist genotyping assays were as described in Vrana et al. (2000). We used modified versions of the earlier Plp1 and Xist assays, which are described below.
Primers used for each respective gene were as follows: CkmF: AAC ACC CAC AAC AAG TTC AAG CTG, CkmR: GTC TGG ATG ACA TCG TCY ASA GTG; EsxF6: GTT TGG TTT CAG AAC AGA AGG, EsxR6: GAT ATT TCC AAA TAA CCT CCA CA; Glra2F: TCA GGA AAA TGC TGC CTC GT, Glra2R: ACA CAC CAT CTC CAT AAA TAC TGT AG; Gpi1F: GTG TGG ACC ACC AGA CAG G, Gpi1R: TTC CGT ATG GGG TGC TGG GTC; Irs4F: TAG GCT TGC TCC CGC TGG A, Irs4R: RAG CCC CGG CCA TTT CCT GAG C; Plp1F3: GGT CCC AGA GAG AAT TAG GCT AGA, Plp1R1: GTA CAC AGG CGC AGC AGA GCA; Prdx4F: TTY YGC GCT YGC KTG GTC ATG GA, Prdx4R: TCT TSG CTT TGC TTA GRT GCA G; XistF2: CTC CCC CTT TAA GAG GGA ACT TGA, XistR2: TAC CCG GAA GTG ACC ACA AT. Annealing temperatures and restriction enzymes used were as follows: Ckm: 57°C, BstUI; Esx1: 52°C, Mbo I ; Glra2: 59°C, HpyCH4IV; Gpi1: 62°C, Aci I; Irs4: 64°C, Bme1580I; Plp1: 58°C, MseI; Prdx4: 52°C, Fnu4HI; Xist: 52°C, BstNI.
Gene expression
RT-PCR was performed on 1.0 μg of DNase I-treated RNA using TaqMan® RT-PCR reagents (Applied Biosystems, Foster City, CA). TaqMan assays for Pw1/ Peg3, Esx1, and an endogenous control, Rpl32, were obtained through the Assays-by-Design service offered by Applied Biosystems after submitting Peromyscus sequence for these genes. In each reaction approximately 75 ng of cDNA was used along with 2.5 μl of 20 × assay mix and TaqMan Universal Mastermix-No UNG (Applied Biosystems) for a total reaction volume of 50 μl. Reactions were done in triplicate in 96-well plates and run on an ABI 7900 real-time PCR instrument. Two or more samples were analyzed for each parental strain and each reciprocal hybrid. Data were analyzed and expression levels calculated as per the manufacturer’s recommendations. Northern analysis was performed by standard methods. Quantitation and visualization was conducted on an Typhoon 9400 (GE Healthcare Bio-Sciences Corp., Piscataway, NJ).
Results
Crosses used to localize Mexl
To refine the area of linkage on the X chromosome, we first bred (bw × po) F1 animals together for multiple generations to create an advanced intercross line (AIL) (Darvasi and Soller 1995). Advanced intercross females were then bred to BW males. We reasoned that such “advanced backcrosses” would enable us to localize the area of the X chromosome responsible for the overgrowth without potential confounding effects of allelic variation at interacting paternal loci.
Our prior analyses were based largely on weight of mature placentas as the result of several factors. First, some PO × BW, PO × AIL, and AIL × BW matings resulted in conceptuses that consisted of only placentas and associated extra-embryonic tissues. Second, those crosses that produced severe overgrowth frequently resulted in maternal lethality. Third, it was very difficult to ensure exact time of embryonic development and thus compare weights of the embryo proper. In contrast, maximum placental weight is reached in Peromyscus at the equivalent stage as in Mus, at approximately 14.5 days of gestation (E14.5).
We had previously undertaken two crosses in which maternal genotype varied. We established X linkage for both adult body weight and placental weight in an F1 female × BW male cross. We then bred a third generation AIL (G3) female × male BW cross resulting in 87 offspring. Placental overgrowth in these embryos had cosegregated most closely with a marker for the gene Plp1 (Mus position = 56 cM). The next closest marker in the initial analysis was that for the Xist gene (Mus position = 42 cM) (Vrana et al. 2000).
Fluorescence in situ hybridization (FISH) analysis has confirmed that the whole Mus X chromosome is syntenic with the Peromyscus X chromosome (Dr. Rachel O’Neill, University of Connecticut, personal. communication) Furthermore, gene order on the X chromosome appears to be generally conserved between the two species (Vrana et al. 2000). We first developed additional markers corresponding to genes on the distal portion of the X chromosome, where we believed Mexl to lie. These markers included genotyping assays for the genes Esx1 (Mus position 57.0 cM), Irs4 (58.0 cM), Prdx4 (67.7 cM), and Glra2 (72.0 cM).
After the previously published work, we next genotyped the G3 × BW offspring with the additional distal X chromosome markers in an attempt to refine the area of phenotypic linkage. In this cross, however, Esx1 genotypes were identical to those of Plp1 for all placentas larger than the parental range. We therefore bred AIL females of greater generation numbers to BW males in the expectation that their offspring would provide greater numbers of recombinant offspring.
Specifically, we utilized both G10 and G13 females in backcrosses to BW males. The two crosses yielded 86 conceptuses of age E14.5 or later. These conceptuses yielded placentas with a spectrum of weights from those within the parental weight range to those exhibiting extreme overgrowth (Table 1). Note that the parental strains show the same mean weight of approximately 0.18 g. We also included eight younger conceptuses whose placental weight already exceeded the maximum weight seen in both parental strains. Thus, we genotyped 94 total offspring of these crosses with the distal X chromosome markers and compared the results with placental weights.
Esx1 maternal genotypes showed the greatest correlation with the largest placentas (Table 2), suggesting this is the closest marker to the Mexl locus. The distribution of maternal Esx1 genotypes by placental weight over all the offspring AIL female × BW male cross is shown in Fig. 1. These data reveal that the PO Esx1 genotypes are also found in normal-sized placentas and that they constitute greater than 92% (87/94) of the total maternal genotypes. We had previously found substantial evidence for allele fixation at multiple autosomal loci in the AIL (Duselis et al. 2005): seven of nine loci tested yielded chi-squared values corresponding to p values of 0.05 or lower. In this case, approximately 31 of the 94 maternal Esx1 alleles are predicted to be of the PO genotype (one third of the original intercross X alleles were a PO genotype). Analysis of this disparity results in a chi-squared score of 101 (p < 0.0005), suggesting that the Esx1 genotypes in the AIL are extremely skewed.
Table 2.
Maternal genotypes of the 20 largest AIL × BW placentas
| Sex | Plac Wt | Xist | Plp1 | Esx1 | Irs4 | Prdx4 | Glra2 |
|---|---|---|---|---|---|---|---|
| M | 0.327 | PO | BW | PO | BW | PO | BW |
| F | 0.328 | PO | PO | PO | PO | PO | BW |
| M | 0.333 | PO | PO | PO | PO | PO | BW |
| F | 0.334 | PO | PO | PO | PO | PO | PO |
| F | 0.338 | PO | PO | PO | PO | PO | BW |
| M | 0.338 | PO | PO | PO | PO | PO | BW |
| F | 0.347 | PO | PO | PO | PO | PO | BW |
| F | 0.357 | PO | PO | PO | BW | PO | PO |
| M | 0.358 | PO | PO | PO | PO | PO | BW |
| F | 0.359 | PO | PO | PO | PO | PO | BW |
| M | 0.362 | BW | PO | PO | BW | BW | BW |
| F | 0.372 | PO | PO | PO | PO | PO | PO |
| M | 0.397 | PO | PO | PO | PO | PO | BW |
| M | 0.419 | PO | PO | PO | PO | PO | BW |
| M | 0.43 | PO | PO | PO | PO | PO | BW |
| M | 0.453 | PO | PO | PO | PO | PO | BW |
| F | 0.456 | PO | PO | PO | PO | PO | BW |
| M | 0.476 | PO | PO | PO | PO | BW | PO |
| M | 0.59 | PO | PO | PO | BW | PO | BW |
| M | 0.673 | PO | PO | PO | PO | PO | BW |
Note that only Esx1 genotypes completely cosegregate with the overgrowth.
Fig. 1.
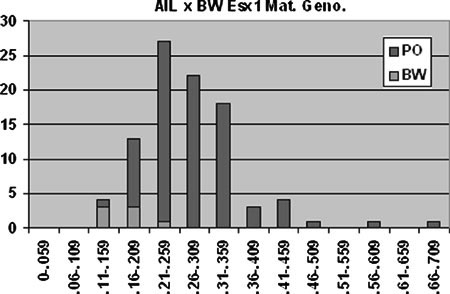
Distribution of the maternal Esx1 alleles in the AIL × BW crosses. Vertical axis represents number of animals, horizontal axis indicates placental weight in grams.
Gene order in this distal portion of the X chromosome appears to be different than that in Mus, where the gene order is (proximal-distal) Xist, Plp1, Esx1, Irs4, Prdx4, Glra2. An analysis of the data in Table 2 using the Map Manager software (Manly and Olson 1999) suggests the gene order in Peromyscus is Prdx4, Xist, Esx1, Plp1, Irs4, with Glra2 either proximal or distal to the other loci.
Crosses used to localize Peal
To localize the paternally expressed gene affecting hybrid growth (Peal), we undertook crosses in which the paternal genome was varied but the maternal genome remained constant. These crosses therefore were PO females crossed with AIL males. Crosses in which the paternal genome is varied but the maternal genome is PO are more difficult to undertake than the reciprocal because of the maternal effect on genomic imprinting control (Duselis et al. 2005). These PO × AIL crosses always result in offspring with some degree of imprinting perturbation and also often in maternal lethality. Resorptions are also common in these crosses, resulting in our harvesting embryos from approximately one quarter of matings.
We obtained 16 litters from PO × AIL (G3 and G13) crosses, resulting in 96 mature placentas for genetic analysis. As in the PO × F1 cross, these offspring displayed mild to severe placental and somatic overgrowth with no obvious sexual dimorphism (Table 1).
We had previously detected linkage of the overgrowth phenotype to the Pw1/Peg3 locus using a PO female × F1 male cross. Pw1/Peg3 is located on Mus Chromosome 7 at 6.5 cM. Mus whole Chromosome 7 FISH probes hybridize entirely to Peromyscus Chromosome 1 (Dawson et al. 1999), suggesting that gene content in this region is conserved. Therefore, we designed genotyping assays for other loci located on Mus proximal Chromosome 7. The new assays include those for Ckm (Mus location = cM 4.5) and Gpi1 (cM 11.0). We genotyped the PO × AIL offspring with these as markers as well as those for Pw1/Peg3, Myod1 (cM 23.5), and H19 (cM 69.0). The latter, however, did not show any genotype-phenotype association (data not shown).
Pw1/Peg3 genotypes exhibited the tightest correlation with placental overgrowth; all placentas above 0.35 g had a BW paternal Pw1/Peg3 genotype. In contrast, at least one of these hyperplastic placentas had a PO genotype at Ckm, Gpi1, and Myod (Table 3). A complete distribution of paternal Pw1/Peg3 genotype over the placental weight range reveals the roughly normal distribution of PO alleles around the parental mean, in contrast to the skewed distribution of the BW alleles (Fig. 2). The allelic content of the cross, however, was not skewed: 57 of the 96 genotyped animals had a BW paternal allele.
Table 3.
Paternal genotypes of the 20 largest PO × AIL placentas
| Plac Wt | Sex | Ckm | Pw1 | Gpi1 | Myod1 |
|---|---|---|---|---|---|
| 0.344 | M | BW | BW | BW | BW |
| 0.346 | F | BW | BW | BW | BW |
| 0.351 | M | BW | BW | BW | BW |
| 0.352 | F | BW | BW | BW | BW |
| 0.354 | M | BW | BW | BW | BW |
| 0.358 | F | BW | BW | BW | BW |
| 0.362 | F | BW | BW | BW | BW |
| 0.385 | F | BW | BW | BW | BW |
| 0.391 | F | BW | BW | BW | BW |
| 0.419 | F | BW | BW | BW | PO |
| 0.421 | M | BW | BW | BW | PO |
| 0.428 | M | BW | BW | BW | PO |
| 0.447 | M | BW | BW | BW | BW |
| 0.451 | M | BW | BW | BW | BW |
| 0.452 | F | BW | BW | BW | BW |
| 0.523 | F | BW | BW | BW | BW |
| 0.561 | F | BW | BW | BW | BW |
| 0.568 | F | BW | BW | BW | BW |
| 0.610 | M | BW | BW | BW | BW |
| 0.612 | M | PO | BW | PO | PO |
Note that only Pw1/Peg genotypes completely cosegregate with the overgrowth.
Fig. 2.
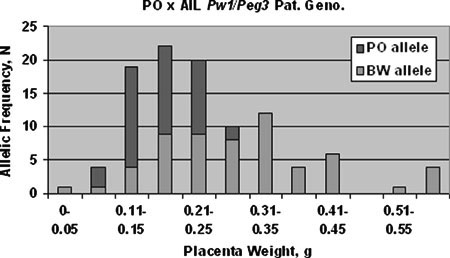
Distribution of the paternal Pw1/Peg3 alleles in the PO × AIL crosses. Vertical axis represents number of animals, horizontal axis indicates placental weight in grams.
To estimate how great an increase in resolution these crosses provided, we compared recombination rates between the F1 and AIL backcrosses. The PO × F1 cross produced four recombinants between Pw1/Peg3 and Myod1 in 109 animals for a distance of 3.7 cM. In contrast, there were 13 recombinants between these two loci in the 103 PO × AIL offspring (12.6 cM). As expected, the majority of the recombinants were found in the later generation AIL crosses. Recombination frequencies between all gene pairs used in mapping Peal are shown in Table 4.
Table 4.
Gene order on Peromyscus Chr 1/Mus Chr 7
| Gene pair | Mus Mb | Pero cM |
|---|---|---|
| Ckm – Pw1/Peg3 | 10.3 | 3.8 |
| Pw1/Peg3 – Gpi1 | 23.4 | 4.9 |
| Gpi1 – Myod1 | 10.8 | 8.7 |
| Ckm – Gpi1 | 13.1 | 1.9 |
| Ckm – Myod1 | 23.9 | 10.7 |
| Pw1/Peg3 – Myod1 | 34.2 | 12.6 |
Distances between gene pairs are shown. House mouse (Mus) is in megabases (Mb) and Peromyscus (Pero) is in centimorgans (cM).
The recombination frequencies confirmed the linkage of these loci on Peromyscus Chromosome 1 as on Mus Chromosome 7. The Mus recombination map, however, differs from the physical map in the relative positions of Ckm and Pw1/Peg3. Our data suggest that the gene order determined for Peromyscus (Pw1/Peg3, Ckm, Gpi1, Myod1) is the same as that for the Mus (as well as Rattus) physical map (http://www.ncbi.nlm.nih.gov/mapview/).
We also tested whether Pw1/Peg3 genotype was also associated with postnatal overgrowth. The PO × F1 backcross produced a limited number of survivors, which were weighed at 45 days after birth. Unlike the conceptuses, the postnatal survivors displayed considerable sexual dimorphism (Fig. 3), with only a subset of males exhibiting overgrowth. Note that all males from this cross have the same sex chromosome genotypes (XPO YPO). Pw1/Peg3 genotypes, however, showed no correlation with body weight (Table 5). In contrast, earlier studies had already demonstrated that Mexl is associated with both prenatal and postnatal growth (Vrana et al. 2000). These data suggest that other autosomal loci may be involved in the overgrowth phenotypes.
Fig. 3.
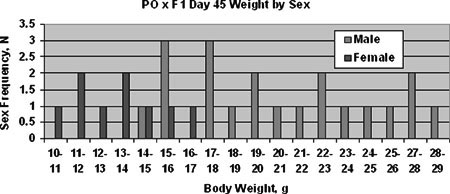
Distribution of sex and weight in the PO × F1 cross at postnatal day 45. Vertical axis represents number of animals, horizontal axis indicates body weight in grams. Parental mean body weights ( ± one standard deviation) are as follows : PO♀ = 15.3 ± 1.2, PO♂ = 12.4 ± 0.4, BW♀ = 15.5 ± 1.6, BW♂ = 17.4 ± 1.4.
Table 5.
Pw1/Peg3 paternal genotypes of the ten largest PO × F1 45-day animals
| Body Wt | Sex | Pw1 |
|---|---|---|
| 20.2 | M | BW |
| 21.9 | M | PO |
| 22.1 | M | PO |
| 22.5 | M | PO |
| 23.2 | M | BW |
| 24 | M | BW |
| 25.9 | M | PO |
| 27.3 | M | PO |
| 27.9 | M | BW |
| 28 | M | BW |
Identification of Mexl and Peal candidate loci
We examined the Mouse Genome Informatics Database (http://www.informatics.jax.org/) for known genes between 50 and 80 cM of the Mus X chromosome. Our criteria for candidate gene identification were (1) loci in the region that show mutant phenotypes similar to those of the Peromyscus hybrids; (2) genes that exhibit great divergence between mouse and human (suggesting rapid evolution that might lead to the interspecific negative interaction); (3) genes thought to be directly involved in growth pathways (e.g., by cell culture assays or paralogy); and (4) genes highly expressed in the placenta.
Another potential criterion for Mexl candidate genes is allelic expression; the genetic analyses suggest it is a maternally expressed gene. Prior work has demonstrated, however, that the paternal X chromosome is preferentially inactivated in Peromyscus extra-embryonic tissues, as it is in Mus (Vrana et al. 2000). That is, the majority of the X chromosome is expressed from the maternal allele in the placenta. Genes in this region fitting one or more of the above criteria are listed in Table 6. Of the 86 genes in this interval, eight fit one and four fit at least two of these criteria. Esx1 alone fulfills three of these criteria (i.e., 1, 2, and 4).
Table 6.
Potential candidate genes for Mexl
| Gene | Function | Map | Criteria |
|---|---|---|---|
| Esx1 | Placental Dev | 57.0 | Plac. Exp., Mutant Growth Pheno. |
| Fgd1 | Nucleotide Exchange | 64.0 | Growth, Plac Exp. |
| Figf | C-fos Induced GF | 70.0 | Growth, Plac. Exp. |
| Irs4 | Insulin Signaling | 58.0 | Mutant Growth Pheno. |
| Magea1-8 | Unknown | 66.1 | Plac. Exp., altered in tumors |
| Ott | Unknown | 62.5 | Plac. Exp. |
| Phex | Proteolysis | 65.4 | Mutant Growth Pheno. |
Map = cM position on Mus X chromosome. Growth = implicated in growth pathways; Plac. Exp. = placental expression; Magea1-8 refers to multiple family members with the corresponding names.
In contrast to the distal X chromosome, proximal Mus Chromosome 7 is gene dense. For example, there are at least 40 distinct genes within 1 cM of Pw1/Peg3. An additional criterion that may be used to identify candidate genes in this region is their allelic expression. That is, our genetic analyses strongly suggest that Peal is normally expressed exclusively from the paternal allele (Vrana et al. 2000). Like other autosomal imprinted domains, only a subset of genes in this region exhibit monoallelic paternal expression. The known paternally expressed genes in this domain include Usp29, Zfp264, and Pw1/Peg3 (http://www.mgu.har.mrc.ac.uk/research/imprinting/).
Comparative genomic analyses, however, suggest that this imprinted domain is rapidly evolving: Domestic cows, humans, and mice show differences in both gene content and imprinting status. For example, Zim2, like Pw1/Peg3, is a paternally expressed zinc-finger gene. The Zim2 gene overlaps the Pw1/Peg3 gene (sharing several small 5′ exons) in humans (Kim et al. 2000). Zim2 is present in Mus but does not share exons with Pw1/Peg3 and is maternally expressed. The Mus genome has another maternally expressed zinc-finger gene, Zim1, which lies between Zim2 and Pw1/Peg3. Both the human and the domestic cow genomes lack Zim1; the latter has an unrelated, nonimprinted gene termed Ast1 in the equivalent position (Kim et al. 2004).
The only imprinted gene in this region to be functionally characterized is Pw1/Peg3. This gene was originally discovered in a screen for novel myogenic regulatory factors. Further biochemical and cell-culture assays have revealed that the gene product acts in the tumor necrosis factor (TNF)/NFkappaB signal transduction pathway and has a key role in p53-mediated apoptosis (Relaix et al. 1998, 2000). The gene-targeting reduction in Pw1/Peg3 product resulted in animals with both placental and somatic growth retardation as well as behavioral defects (Curley et al. 2004; Li et al. 1999). Thus, one might predict that overexpression of the gene resulting from the LOI seen in the PO × BW hybrids would lead to overgrowth.
Another locus that maps to the equivalent region in humans, 19q13.4, also affects placental growth (Panichkul et al. 2005). This locus influences susceptibility to biparental complete hydatidiform moles (BiCHM), which are characterized by the presence of hyperplastic trophoblast villi and lack of a fetus. Hydatidiform moles typically have either no maternal genome or have two paternal genomes (e.g., due to fertilization by two sperm) (Li et al. 2002; Szulman 1987). Recent studies, however, have confirmed the existence of hydatidiform moles that have normal biparental genomic contributions (BiCHM). The BiCHM tissues exhibit altered DNA methylation suggestive of LOI (Judson et al. 2002).
While the familial BiCHM susceptibility locus has been localized to the Pw1/Peg3 domain, it does not correlate with coding region polymorphisms of the gene. The gene NALP7, known to be involved in apoptotic and inflammatory pathways, has emerged as the primary candidate for those BiCHM whose susceptibility locus maps to Chromosome 19 (Murdoch et al. 2006). The Mus genome, however, apparently lacks an ortholog in this region.
While we do not yet know the structure of this domain in Peromyscus, Northern analysis suggests that there are no genes sharing exons with Pw1/Peg3. We hybridized placental and embryonic RNA with multiple probes derived from regions equivalent to the Mus upstream exons and the large (∼5 kb) final exon. All tests revealed only the approximately 9-kb transcript also observed in Mus (data not shown).
Expression analysis of candidate loci
Upon identifying Esx1 and Pw1/Peg3 as candidate genes for Mexl and Peal (respectively), we asked whether expression levels varied between the two species and if perturbations occurred in the hybrid offspring. We tested expression levels in both genes via Northern analysis and quantitative PCR analysis.
Esx1 expression has been shown to be confined to the placenta and testis in Mus; Northern analysis of PO and BW mid-late gestation embryos suggests that this is also true in Peromyscus. However, we included hybrid embryos in the quantitative PCR assays in the event spatial expression was perturbed. Esx1 did not exhibit discernible expression differences between the two parental strains nor between the oversized hybrids and the parental strains (Fig. 4). In contrast, the growth-retarded bw × po placentas exhibit substantial upregulation of the Esx1 transcript. Note that the complete loss of Esx1 results in an enlarged placenta (Li and Behringer 1998).
Fig. 4.
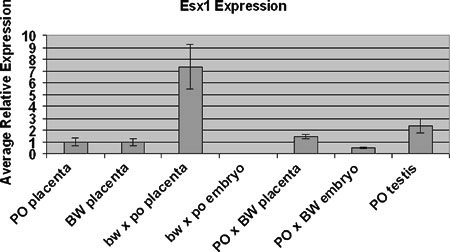
Quantitative PCR analysis of Esx1 expression. Each indicated genotype-tissue type combination represents multiple samples, with the exception of testis. Standard error bars are indicated.
It is unclear whether Esx1 is a viable candidate gene for the genetic incompatibilities underlying the hybrid growth retardation. While preliminary evidence suggests the involvement of the X chromosome in the genetics of the bw × po hybrid phenotypes, it is not yet clear whether the same region is involved. Esx1 is predominantly expressed in the labyrinth region of the placenta. The other major layer of the muroid placenta, the spongiotrophoblast or junctional zone, is greatly reduced in the bw × po placentas (Rogers and Dawson 1970; Vrana et al. 2001). Therefore, the increased Esx1 expression in bw × po may be a consequence of a relative overabundance of the expressing cells.
The H19 gene is also predominantly transcribed in the labyrinthine portion of the Peromyscus placenta (Vrana et al. 2001). However, total H19 expression levels in the bw × po placenta are no higher than that seen in either parental strain (Vrana et al. 1998). Thus, it is possible that the increased Esx1 expression plays a causal role in the growth-retarded hybrid placentas.
We also tested Esx1 allelic usage in the Peromyscus hybrids. Like other X-linked genes, Esx1 was expressed exclusively from the maternal allele in extra-embryonic tissues (data not shown). In contrast, Pw1/Peg3 has been shown to lose imprinting in the PO × BW cross (Duselis et al. 2005; Vrana et al. 1998). That is, the gene is expressed from both alleles in the oversized hybrids. Corresponding to this biallelic expression, we observed increased expression levels in PO × BW conceptuses, particularly in the embryo proper (Fig. 5). Surprisingly, Pw1/Peg3 also showed increased expression in the bw × po embryos, despite maintaining strict paternal expression.
Fig. 5.
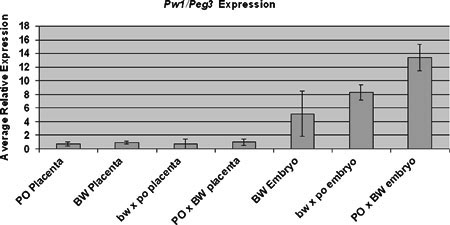
Quantitative PCR analysis of Pw1/Peg3 expression. Each indicated genotype-tissue type combination represents multiple samples, with the exception of testis. Standard error bars are indicated.
Discussion
These Peromyscus species and their hybrids offer a novel system for studying the role of natural genetic variation on epigenetic regulation, reproductive isolation, growth, and development. Here we have refined the areas of the genome containing two loci (Mexl and Peal) contributing to the hybrid parent-of-origin effects. Peal remains unbounded on one side. The combination of tight linkage to Pw1/Peg3 in combination with requirement for paternal expression, however, will likely result in few other Peal candidate loci. As most X-linked loci are maternally expressed in rodent placentas, it would be difficult to identify Mexl candidates on the basis of allelic expression. Mexl, however, appears to be bounded by Plp1 and Esx1 (Table 2). These two loci lie only about 300 kb apart in Mus (http://www.ensembl.org/Mus_musculus). Thus, a comprehensive survey of this region in Peromyscus should not be difficult.
We have identified candidate genes for both loci, Esx1 for Mexl and Pw1/Peg3 for Peal. Genotypes of both candidate loci are completely concordant with placental hypertrophy in their respective backcrosses. While both genes show equivalent expression levels in mature placentas of the parental strains, this does not rule out interspecific regulatory changes. For example, the genes may show different temporal and/or spatial expression patterns.
Despite the apparent requirement of these alleles for hypertrophy, those particular genotypes (PO maternal allele of Esx1, BW paternal allele of Pw1/Peg3) did not always result in overgrowth. Our approach of crossing AIL animals to a parental strain rules out incomplete penetrance because of the necessity of genotypes at interacting loci inherited from the other parent.
It seems likely, however, that other alleles from the varying parental genome may act as modifiers to the phenotypes. For example, the lack of correlation of Pw1/Peg3 genotypes with postnatal overgrowth in the PO × F1 cross suggests the involvement of other loci contributing to the hybrid overgrowth. Furthermore, the PO × F1 placental analysis revealed a class of mildly overgrown placentas that were also not dependent on Pw1/Peg3 genotype (Vrana et al. 2000). Another unidentified locus affecting postnatal overgrowth in the F1 female × BW backcross has been linked to the autosomal platinum coat-color allele, which does not exhibit linkage to Pw1/Peg3 (W. Dawson, J. Rice, J. Weston, C. Ramsdell, and M. Dewey; submitted).
The Pw1/Peg3 region is gene dense, potentially rendering further genetic tests of Peal difficult. A first approach would be to characterize the genes in the region and their allelic expression, i.e., nonpaternally expressed genes could be ruled out. A systematic study of this domain has not yet been performed in any species. Furthermore, comparative studies to date suggest one cannot assume that imprinting status in this region necessarily applies to other taxa (Morison et al. 2005).
In contrast, the paucity of genes on the distal X chromosome of muroid rodents examined to date makes potential testing of candidate loci more feasible. While Esx1 looks promising as a candidate, it is unclear how its gene product might regulate postnatal growth, given its lack of embryonic and postnatal expression (save testis) (Li and Behringer 1998; Li et al. 1997).
Crosses between the mixed genetic background Mus strain C57BL/6 (B6) and Mus spretus also yield reciprocal parent-of-origin effects on placental size. The Mus crosses, however, do not yield significant reciprocal effects on embryo or postnatal growth. Similar to the Peromyscus crosses, the Mus hybrid placental growth effects show genetic linkage to the X chromosome and altered Esx1 expression (Zechner et al. 2002). No genetic role for any autosomal locus has yet been found in the Mus hybrids (Zechner et al. 1997).
The greater inquiries underlying this work are whether these allelic variants of imprinted genes and imprinting regulatory genes have undergone positive selection, whether similar changes underlie other cases of reproductive isolation and human disease, and what the commonalities are in the biological roles of imprinted genes.
It is clear that a major role of imprinted gene products is to modulate prenatal growth. The placenta, a key endocrine organ, expresses many of these loci (Reik et al. 2003). Eutherian placentas, however, vary tremendously in overall morphology, cell types, and gene expression (Carter and Enders 2004; Cross et al. 2003). Numerous associations between altered imprinted gene expression and placental phenotypes suggest at least a role for imprinted genes in this aspect of mammalian diversity. Evidence suggests roles for imprinted genes in a number of human growth and placental disorders (Fisher and Hodges 2003; Reik and Walter 2001). Included in the latter group are the two most common, preeclampsia (Oudejans et al. 2004) and gestational diabetes (Reik et al. 2003). Thus, it seems likely that natural variation at imprinted loci can contribute to mammalian speciation. This work represents a significant advance in isolating such variation.
An opportunity exists to generally test whether Mexl and Peal play a role in reproductive isolation in other populations. The Peromyscus maniculatus species complex contains a number of partially isolated populations. Furthermore, preliminary studies suggest parent-of-origin effects in hybrids between several of these populations (Dice 1949; Liu 1954). Because very little is known about genetic and epigenetic variation at imprinted loci in any mammalian species (Vrana 2006), the present work provides a preliminary framework to test hypotheses about this aspect of mammalian biology.
Acknowledgments
This work was supported by a grant from the American Cancer Society (RSG-03-070-01-MGO) to P.B.V. The authors thank Shirley Tilghman, Mike Dewey, and Cliff Ramsdell for comments and discussion.
Footnotes
Maria Loschiavo and Quang K. Nguyen contributed equally to this work.
References
- Carter AM, Enders AC (2004) Comparative aspects of trophoblast development and placentation. Reprod Biol Endocrinol 2, 46 [DOI] [PMC free article] [PubMed]
- Cross JC, Baczyk D, Dobric N, Hemberger M, Hughes M, et al. (2003) Genes, development and evolution of the placenta. Placenta 24, 123–130 [DOI] [PubMed]
- Curley JP, Barton S, Surani A, Keverne EB (2004) Coadaptation in mother and infant regulated by a paternally expressed imprinted gene. Proc Biol Sci 271, 1303–1309 [DOI] [PMC free article] [PubMed]
- Darvasi A, Soller M (1995) Advanced intercross lines, an experimental population for fine genetic mapping. Genetics 141, 1199–1207 [DOI] [PMC free article] [PubMed]
- Dawson WD (1965) Fertility and size inheritance in a Peromyscus species cross. Evolution 19, 44–55 [DOI]
- Dawson WD, Sagedy MN, En-yu L, Kass DH, Crossland JP (1993) Growth regulation in Peromyscus species hybrids: a test for mitochondrial-nuclear genomic interaction. Growth Dev Aging 57, 121-133 [PubMed]
- Dawson WD, Young SR, Wang Z, Liu LW, Greenbaum IF, et al. (1999) Mus and Peromyscus chromosome homology established by FISH with three mouse paint probes. Mamm Genome 10, 730–733 [DOI] [PubMed]
- Dewey MJ, Dawson WD (2001) Deer mice: “The Drosophila of North American mammalogy.” Genesis 29, 105–109 [DOI] [PubMed]
- Dice LR (1949) Variation of Peromyscus maniculatus in parts of Western Washington and adjacent Oregon. Contrib Lab Vert Biol 44, 1–34
- Duselis AR, Wiley CD, O’Neill MJ, Vrana PB (2005) Genetic evidence for a maternal effect locus controlling genomic imprinting and growth. Genesis 43, 155–165 [DOI] [PubMed]
- Fisher RA, Hodges MD (2003) Genomic imprinting in gestational trophoblastic disease—a review. Placenta Suppl A, S111–1118 [DOI] [PubMed]
- Judson H, Hayward BE, Sheridan E, Bonthron DT (2002) A global disorder of imprinting in the human female germ line. Nature 416, 539–542 [DOI] [PubMed]
- Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD (2004) The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat Genet 36, 642–646 [DOI] [PubMed]
- Kim J, Bergmann A, Lucas S, Stone R, Stubbs L (2004) Lineage-specific imprinting and evolution of the zinc-finger gene ZIM2. Genomics 84, 47–58 [DOI] [PubMed]
- Kim J, Bergmann A, Stubbs L (2000) Exon sharing of a novel human zinc-finger gene, ZIM2, and paternally expressed gene 3 (PEG3). Genomics 64, 114–118 [DOI] [PubMed]
- Li HW, Tsao SW, Cheung ANY (2002) Current understandings of the molecular genetics of gestational trophoblastic diseases. Placenta 23, 20–31 [DOI] [PubMed]
- Li L, Keverne EB, Aparicio SA, Ishino F, Barton SC, et al. (1999) Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science 284, 330–333 [DOI] [PubMed]
- Li Y, Behringer R (1998) Esx1 is an X-chromosome-imprinted regulator of placental development and fetal growth. Nat Genet 20, 309–311 [DOI] [PubMed]
- Li Y, Lemaire P, Behringer RR (1997) Esx1, a novel X chromosome-linked homeobox gene expressed in mouse extraembryonic tissues and male germ cells. Dev Biol 188, 85–95 [DOI] [PubMed]
- Liu TT (1954) Hybridization between Peromyscus maniculatus oreas and P. m. gracilis. J Mamm 35, 448–449 [DOI]
- Manly KF, Olson JM (1999) Overview of QTL mapping software and introduction to map manager QT. Mamm Genome 10, 327–334 [DOI] [PubMed]
- Moore T, McLellan A, Wynne F, Dockery P (2005) Explaining the X-linkage bias of placentally expressed genes. Nat Genet 37, 3 [DOI] [PubMed]
- Morison IM, Ramsay JP, Spencer HG (2005) A census of mammalian imprinting. Trends Genet 21, 457–465 [DOI] [PubMed]
- Murdoch S, Djuric U, Mazhar B, Seoud M, Khan R, et al. (2006) Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet 38, 300–302 [DOI] [PubMed]
- Oudejans CB, Mulders J, Lachmeijer AM, van Dijk M, Konst AA, et al. (2004) The parent-of-origin effect of 10q22 in pre-eclamptic females coincides with two regions clustered for genes with down-regulated expression in androgenetic placentas. Mol Hum Reprod 10, 589–598 [DOI] [PubMed]
- Panichkul PC, Al-Hussaini TK, Sierra R, Kashork CD, Popek EJ, et al. (2005) Recurrent biparental hydatidiform mole: additional evidence for a 1.1-Mb locus in 19q13.4 and candidate gene analysis. J Soc Gynecol Investig 12, 376–383 [DOI] [PubMed]
- Reik W, Walter J (2001) Genomic imprinting: parental influence on the genome. Nat Rev Genet 2, 21–32 [DOI] [PubMed]
- Reik W, Constancia M, Fowden A, Anderson N, Dean W, et al. (2003) Regulation of supply and demand for maternal nutrients in mammals by imprinted genes. J Physiol 547, 35–44 [DOI] [PMC free article] [PubMed]
- Relaix F, Wei XJ, Wu X, Sassoon DA (1998) Peg3/Pw1 is an imprinted gene involved in the TNF-NFkappaB signal transduction pathway. Nat Genet 18, 287–291 [DOI] [PubMed]
- Relaix F, Wei X, Li W, Pan J, Lin Y, et al. (2000) Pw1/Peg3 is a potential cell death mediator and cooperates with Siah1a in p53-mediated apoptosis. Proc Natl Acad Sci U S A 97, 2105–2110 [DOI] [PMC free article] [PubMed]
- Rogers JF, Dawson WD (1970) Foetal and placental size in a Peromyscus species cross. J Reprod Fertil 21, 255–262 [DOI] [PubMed]
- Szulman AE (1987) Clinicopathologic features of partial hydatidiform mole. J Reprod Med 32, 640–643 [PubMed]
- Takagi N, Sasaki M (1975) Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256, 640–642 [DOI] [PubMed]
- Vrana PB (2006) Genomic imprinting as a force in mammalian diversity. J Mammal in press
- Vrana PB, Guan XJ, Ingram RS, Tilghman SM (1998) Genomic imprinting is disrupted in interspecific Peromyscus hybrids. Nat Genet 20, 362–365 [DOI] [PubMed]
- Vrana PB, Fossella JA, Matteson P, del Rio T, O’Neill MJ, et al. (2000) Genetic and epigenetic incompatibilities underlie hybrid dysgenesis in Peromyscus. Nat Genet 25, 120–124 [DOI] [PubMed]
- Vrana PB, Matteson PG, Schmidt JV, Ingram RS, Joyce A, et al. (2001) Genomic imprinting of a placental lactogen in Peromyscus. Dev Genes Evol 211, 523-532 [DOI] [PubMed]
- Xue F, Tian XC, Du F, Kubota C, Taneja M, et al. (2002) Aberrant patterns of X chromosome inactivation in bovine clones. Nat Genet 31, 216-220 [DOI] [PubMed]
- Zechner U, Reule M, Burgoyne PS, Schubert A, Orth A, et al. (1997) Paternal transmission of X-linked placental dysplasia in mouse interspecific hybrids. Genetics 146, 1399-1405 [DOI] [PMC free article] [PubMed]
- Zechner U, Hemberger M, Constancia M, Orth A, Dragatsis I, et al. (2002) Proliferation and growth factor expression in abnormally enlarged placentas of mouse interspecific hybrids. Dev Dyn 224, 125–134 [DOI] [PubMed]


