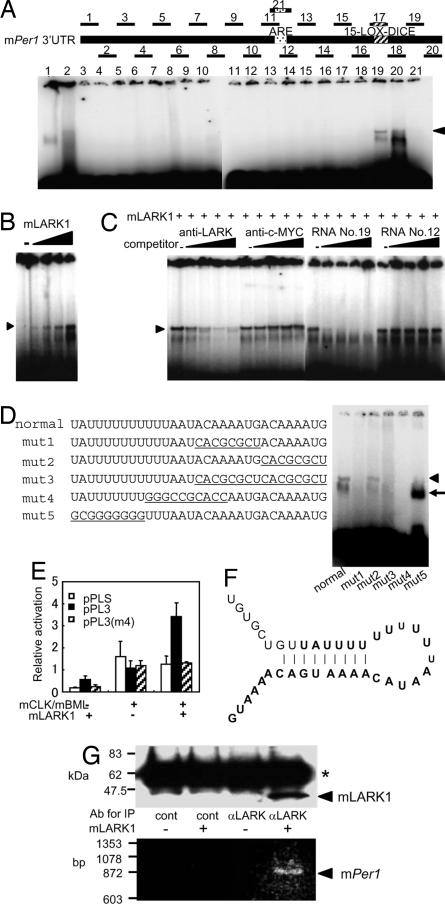
Fig. 4.
Direct interaction between mLark1 and mPer1 3′ UTR. (A) RMSA of His-mLARK1 and RNAs derived from mPer1 3′ UTR is shown. The numbers correspond to RNA oligonucleotides that span the entire mPer1 3′ UTR as diagrammed at the top (the sequences are listed in SI Table 2). The position of the previously identified ARE and LOX-DICE elements are marked (18). Arrowhead denotes the LARK-specific retarded band. (B) Dose–response relationship between His-mLARK1 and RNA 19 is shown. Shown is an RMSA as in A, but using only RNA 19 as probe and with increasing doses of His-mLARK1 as indicated. (C) Competitive analyses verify specificity of interaction between mLARK1 and RNA. RMSAs were done as in B but with the addition of increasing amounts of antibody or cold RNA competitors as indicated. (D) RMSA using normal and mutated versions of RNA 19 is shown. Underlined nucleotides are mutated nucleotides. The arrowhead indicates the RNA bands retarded by mLARK1, and the arrow indicates a nonspecific band. (E) LARK1 only activates reporter gene expression when the LARK binding site is intact. Reporter analyses were done as in Fig. 2, except an additional reporter construct was included in which the mPer1 3′ UTR contains the mutations shown in mut4 (D) [pPL3(m4)]. (F) The predicted RNA secondary structure is shown. Bold type indicates nucleotides corresponding to RNA fragment 19. (G) The direct interaction between LARK1 protein and Per1 mRNA is shown. Extracts from NIH 3T3 cells with either overexpressed mLARK1 or control plasmid were immunoprecipitated with either anti-LARK or control antibody. Immunoprecipitates were analyzed by Western blot using LARK antibody (Upper) or RT-PCR for mPer1 (Lower). Asterisk indicates the IgG heavy chain from the immunoprecipitating antibody.