The gospel of Matthew (chapter 8, verse 14) reports that “Peter's mother-in-law was sick of the fever,” and many commentaries think that this close relative of the apostle living in the Galilee ≈2,000 years ago was suffering from malaria. The main characteristic of malaria is fever, and periodic fevers have been reported even >3,000 years ago in early Chinese, Chaldean, Hindu, Egyptian, and Greek writings (1, 2). It is likely that some, although surely not all of them, were malarial. Major discoveries in the late 19th century by Laveran (identifying Plasmodium as the pathogen causing malaria) and by Ross (describing the life cycle of the parasite and mosquitoes as the vector) led to advances in partially controlling this devastating disease. It is, however, still affecting ≈400 million people worldwide, and its complications, such as cerebral malaria caused by Plasmodium falciparum, still have a high mortality rate (3). The pathophysiology of the host–parasite interaction and, particularly, the mechanism of fever induction in malaria until recently has been far from being understood. However, knowledge of the malaria pathology is urgently needed to potentially develop novel intervention strategies to efficiently reduce the high disease burden causing >2 million deaths per year. In this issue of PNAS, Parroche et al. (4) report on a novel mechanism that the host uses to recognize Plasmodium DNA via the Toll-like receptor (TLR) 9, which may be a key step for inducing fever during this disease. These findings reveal an important mechanism of disease pathophysiology that may also apply to other microbial diseases.
The protein family of TLRs has been discovered recently and was functionally analyzed by scientists during the last 8 years. Together with the intracellular nod-like receptors (NLRs), they are key recognition molecules for microorganisms including viruses, bacteria, and parasites (5). Engagement of most of the TLRs triggers a signaling pathway leading to the translocation of the transcription factor NF-κB into the nucleus, followed by the subsequent release of proinflammatory cytokines. Apart from other activities, many of these factors induce the activation of the inducible cyclooxygenase (COX)-2, which up-regulates prostaglandin synthesis, changing the set-point of the thermoregulatory center of the host leading to fever (6). One group of the 11 members of the TLR family is located on the cell surface, recognizing cell wall components of microorganisms, whereas the other group is intracellularly expressed and recognizes nucleic acids, such as single-stranded RNA (TLR7 and TLR8), double-stranded RNA (TLR3), or DNA (TLR9). The innate immune system is the first line of defense potentially controlling microorganisms at an early stage by inducing inflammation and fever. Malaria is a disease leading to an extremely high pathogen burden on one hand and a typically strong febrile response on the other hand. It thus was of great interest to identify the specific molecular mechanisms of the early immune recognition of Plasmodium to understand disease pathophysiology and to potentially develop more successful vaccination and/or antiparasitic strategies.
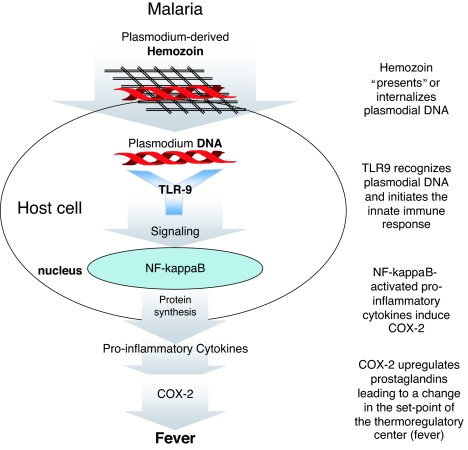
Protozoa, such as P. falciparum causing the most severe form of malaria, contain a glycosylphosphatidylinositol (GPI) anchor on the cell surface, and this molecule is recognized by TLR2 and TLR4 (reviewed in ref. 7). For malaria, however, the GPI anchor–TLR2 interaction is comparably weak; thus, other interactions with the TLR system have been suggested. Recently, Coban et al. (8) showed that hemozoin, the malaria pigment found in large quantities in macrophages during malaria, can stimulate the host via TLR9. These results were quite surprising because TLR9 had been convincingly described as a receptor for DNA, mainly of unmethylated, CpG-containing DNA, frequently found in bacteria. In this publication, “contaminating” DNA was ruled out by DNase treatment and the failure to result in a reduction in TLR9-stimulating capacity of the hemozoin preparations. Although this discrepancy remains unclear, the work of Parroche et al. (4) shows that it is plasmodial DNA instead that stimulates TLR9 of the host and that this activity is clearly DNase-sensitive. The Golenbock group (4), in addition, convincingly shows that hemozoin plays a specific role in presenting the DNA to the intracellular TLR9; however, it cannot stimulate the innate immune system by itself (Fig. 1).
Fig. 1.
Potential mechanism of malaria-induced fever. Parroche et al. (4) show that hemozoin contains plasmodial DNA and that it “presents” or internalizes DNA. Plasmodial DNA then intracellularly interacts with TLR9, initiating signal transduction leading to the release of proinflammatory cytokines via NF-κB activation. These cytokines induce COX-2-up-regulating prostaglandins, which subsequently leads to the induction of fever.
The principle of microbial DNA being recognized by the host is fascinating, and the work presented here gives a surprising new example of this defense mechanism. An interesting paradox, however, is that although TLR9 is known for recognizing GC-rich areas typically found, e.g., in bacterial DNA, plasmodial DNA has one of the highest AT contents known in nature. This AT-richness has even been exploited for therapeutic strategies against malaria (9) and could, in my opinion, potentially explain why malaria parasites remain unrecognized by the innate immune system, leading to a high pathogen burden found only in malaria. On the other hand, the authors of this study (4) present evidence that plasmodial DNA, although overall extremely AT-rich, has small GC-rich regions that are able to stimulate TLR9 efficiently. Although a mere blocking of the (“physiological”) fever responses of the host most likely will not lead to new therapeutic success against this disease, this interaction of plasmodial DNA with TLR9 could lead to new strategies for malaria vaccines and immunomodulation because TLR9-engagement is known for its strong adjuvant potential, which currently is tested for tumor-vaccination strategies and many other areas of immunomodulation (10). TLR interaction of Plasmodium may also be involved in a phenomenon called malaria tolerance or semiimmunity (11). Here, individuals, after having recovered from malaria disease, upon subsequent encounters with the parasite, present with parasitemia without becoming severely ill. Based on the findings of Parroche et al. (4), plasmodial DNA–TLR9 interaction could represent a mechanism for this “malaria tolerance,” which, if true, could also be exploited therapeutically in the future. The data obtained by Parroche et al. could finally give an additional explanation for the mechanism of action of the antimalaria drug chloroquin: This substance is known to interfere with both hemozoin formation and nuclease activity on one side and intracellular TLR9 processing by blocking endosomal acidification on the other (12). If this interference with the DNA–TLR9 interaction proved to be the major mechanism of action of this efficient antimalarial drug, it would show that the mechanism described is of central importance for malaria pathology. In an animal model, for example, it has been shown previously that chloroquin completely protects from cytokine release induced by CpG oligonucleotide–TLR9 interaction (13).
Very recently, it was shown in a mouse model of cerebral malaria employing Plasmodium berghei that deficiency in both TLR2 and TLR9 protects from the lethal outcome of infection (14). Although this model may not be able to perfectly mimic human malaria, it supports the notion that blocking of TLR9 could potentially modulate the disease in humans. TLR9-blocking substances such as specific oligonucleotides are currently being developed and tested by several companies (15) and, potentially, should be tested as antimalaria strategy as well.
Analyzing the genome of patients and relating it to disease susceptibility in recent years has revealed individual genetic variations of the TLR system to influence disease susceptibility substantially (16). We recently published results from genotyping studies originating from Ghana, Africa, supporting the role of TLR9 during malaria: It is known that pregnant women developing malaria carry a particularly high risk for both maternal and child complications, causing the death of up to 10,000 pregnant women and 200,000 newborns annually (17). It is also known that in the placenta, extremely large quantities of hemozoin are accumulating during this disease. We found that in women with malaria during pregnancy, certain genetic variants of TLR9 predispose for low birth weight of the child (18). In contrast, overall susceptibility to malaria in this study did not seem to associate with the genetic TLR9 variants tested here; such a correlation, however, was found for TLR4 SNPs, whereas otherwise frequent TLR2 SNPs, surprisingly, were completely absent (19).
Genetics of the Anopheles gambiae mosquito, the major vector for P. falciparum, recently gave further support for the notion that the TLR system plays a crucial role in disease development: Resistance against P. falciparum as a prerequisite of this insect to be the vector of Plasmodium could be linked to a genetic area within A. gambiae coding for two novel leucine-rich-repeat (LRR)-containing proteins termed APL1 and APL2 (20). These proteins very likely represent a defense system equivalent to the TLR/NLR system of mammals (also typically containing LRRs), and a genetic defect that is crucial for resistance of the mosquito against Plasmodium supports the central role of this host system for malaria defense.
In summary, the work of Parroche et al. elucidates an important step in malaria pathophysiology and, particularly, resolves a long-time mystery: Why and how does malaria cause fever? It also corrects previous findings claiming that hemozoin is a direct TLR9 stimulus and refines them by showing that hemozoin itself is important for presenting the DNA to TLR9 but does not stimulate the receptor. Although it is too early to predict how these findings will influence the development of future malaria treatment options, it is likely that it will open new pathways of interference with the malaria fever reaction, and this may influence the course of disease. The biblical story quoted earlier ends with a miraculous healing of the fever of Peter's mother-in-law. Because we may not want to rely only on miracles today when it comes to this still-devastating disease, we may, in the future, be able to develop scientifically founded ways to interfere with the fever based on the findings presented here and, hopefully, gain new prophylactic and treatment options for an old but still too up-to-date disease.
Abbreviation
- TLR
Toll-like receptor.
Footnotes
The author declares no conflict of interest.
See companion article on page 1918.
References
- 1.Wyler D. In: Infectious Diseases. Gorbach S, Bartlett J, Blacklow N, editors. Philadelphia: Saunders; 1998. pp. 2407–2420. [Google Scholar]
- 2.Retief F, Cilliers L. S Afr Med J 96. 2006;684:686–688. [PubMed] [Google Scholar]
- 3.Guinovart C, Navia MM, Tanner M, Alonso PL. Curr Mol Med. 2006;6:137–140. doi: 10.2174/156652406776055131. [DOI] [PubMed] [Google Scholar]
- 4.Parroche P, Lauw FN, Goutagny N, Latz E, Monks BG, Visintin A, Halmen KA, Lamphier M, Olivier M, Bartholomeu DC, et al. Proc Natl Acad Sci USA. 2006;104:1918–1924. doi: 10.1073/pnas.0608745104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawai T, Akira S. Curr Opin Immunol. 2005;17:338–344. doi: 10.1016/j.coi.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Ball HJ, MacDougall HG, McGregor IS, Hunt NH. J Infect Dis. 2004;189:751–758. doi: 10.1086/381503. [DOI] [PubMed] [Google Scholar]
- 7.Gazzinelli RT, Denkers EY. Nat Rev Immunol. 2006;6:895–906. doi: 10.1038/nri1978. [DOI] [PubMed] [Google Scholar]
- 8.Coban C, Ishii KJ, Kawai T, Hemmi H, Sato S, Uematsu S, Yamamoto M, Takeuchi O, Itagaki S, Kumar N, et al. J Exp Med. 2005;201:19–25. doi: 10.1084/jem.20041836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ginsburg H, Nissani E, Krugliak M, Williamson DH. Mol Biochem Parasitol. 1993;58:7–15. doi: 10.1016/0166-6851(93)90085-c. [DOI] [PubMed] [Google Scholar]
- 10.Krieg AM. Nat Rev Drug Discov. 2006;5:471–484. doi: 10.1038/nrd2059. [DOI] [PubMed] [Google Scholar]
- 11.Boutlis CS, Yeo TW, Anstey NM. Trends Parasitol. 2006;22:371–377. doi: 10.1016/j.pt.2006.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim EJ, Lee SH, Lee JG, Chin BR, Bae YS, Kim JR, Lee CH, Baek SH. FEBS Lett. 2006;580:4533–4538. doi: 10.1016/j.febslet.2006.06.100. [DOI] [PubMed] [Google Scholar]
- 13.Hong Z, Jiang Z, Liangxi W, Guofu D, Ping L, Yongling L, Wendong P, Minghai W. Int Immunopharmacol. 2004;4:223–234. doi: 10.1016/j.intimp.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Coban C, Ishii KJ, Uematsu S, Arisue N, Sato S, Yamamoto M, Kawai T, Takeuchi O, Hisaeda H, Horii T, Akira S. Int Immunol. 2007;19:67–79. doi: 10.1093/intimm/dxl123. [DOI] [PubMed] [Google Scholar]
- 15.Rezaei N. Int Immunopharmacol. 2006;6:863–869. doi: 10.1016/j.intimp.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Schroder NW, Schumann RR. Lancet Infect Dis. 2005;5:156–164. doi: 10.1016/S1473-3099(05)01308-3. [DOI] [PubMed] [Google Scholar]
- 17.Becker K, Tilley L, Vennerstrom JL, Roberts D, Rogerson S, Ginsburg H. Int J Parasitol. 2004;34:163–189. doi: 10.1016/j.ijpara.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 18.Mockenhaupt FP, Hamann L, von Gaertner C, Bedu-Addo G, von Kleinsorgen C, Schumann RR, Bienzle U. J Infect Dis. 2006;194:184–188. doi: 10.1086/505152. [DOI] [PubMed] [Google Scholar]
- 19.Mockenhaupt FP, Cramer JP, Hamann L, Stegemann MS, Eckert J, Oh NR, Otchwemah RN, Dietz E, Ehrhardt S, Schroder NW, et al. Proc Natl Acad Sci USA. 2006;103:177–182. doi: 10.1073/pnas.0506803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riehle MM, Markianos K, Niare O, Xu J, Li J, Toure AM, Podiougou B, Oduol F, Diawara S, Diallo M, et al. Science. 2006;312:577–579. doi: 10.1126/science.1124153. [DOI] [PubMed] [Google Scholar]