Figure 1.
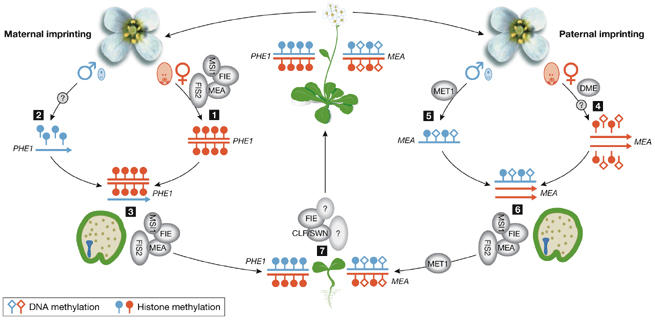
Model of molecular mechanisms controlling maternal and paternal imprinting in Arabidopsis. The maternally imprinted gene PHERES 1 (PHE1; red) is repressed in the female gametophyte by the FERTILIZATION INDEPENDENT SEED (FIS) Polycomb group (PcG) complex, which contains MEDEA (MEA), FERTILIZATION INDEPENDENT ENDOSPERM (FIE), FIS2 and MULTI-COPY SUPPRESSOR OF IRA1 (MSI1) and applies methylation on histone H3 lysine 27 (H3Lys27; 1). From the active paternal PHE1 allele (blue) H3Lys27 methylation needs to be removed, but it is not known whether this is an active or passive process (2). Repression of the maternal PHE1 allele in the endosperm is maintained by the FIS complex (3). In the central cell of the female gametophyte, DEMETER (DME) actively erases DNA methylation from the maternal allele (red) of the paternally imprinted gene MEA (4). It is not known whether repressive H3Lys27 methylation is actively erased from the maternal MEA allele as well. The paternal MEA allele (blue) is methylated by DNA METHYLTRANSFERASE 1 (MET1). Whether repressive histone methylation is maintained on the paternal allele is unknown (5). Repression of the paternal MEA allele in the endosperm is maintained by the FIS complex (6). In vegetative tissues, the silent state of both genes, PHE1 and MEA is maintained by H3Lys27 methylation (7), which depends on PcG complexes containing CURLY LEAF (CLF), SWINGER (SWN) and FIE.
