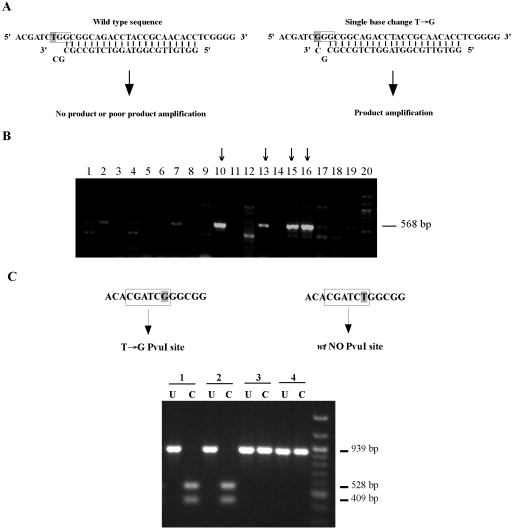
Fig. 5.
Construction of a point mutant in TM4 Tmp mt3. To construct a point mutation in the shuttle phasmid phAE87, two complementary single-strand oligonucleotides (100-mer) containing a T→G substitution that changes a TGG for a GGG codon (W853G) in gene 17 were electroporated into an E. coli recombineering strain and pools of recovered cells screened first by allele-specific PCR amplification (MAMA-PCR) (Cha et al., 1992), and then by diagnostic restriction digestion. A. Schematic representation of MAMA-PCR. A detection primer (reverse primer) with a two-base mismatch at the 3′ does not efficiently amplify the wild-type sequence under the PCR conditions used (see Experimental procedures), although the same primer anneals with the mutated sequence and effectively amplifies a 568 bp product. B. PCR to identify positive clones after recombineering. Pools of recombineering products (approximately five cells/pool) were analysed by MAMA-PCR and the products analysed by agarose gel electrophoresis. Lanes 2–19 contain representative pools, and those containing phasmids with the desired single base substitution T→G (phAE8717W853G) are indicated with an arrow. Lane 1 contains parent control DNA and lane 20 is a DNA marker. Of the 93 pools tested, 25 contained at least one mutant derivative, corresponding to a frequency of approximately one recombinant per 20 electroporated cells. C. Diagnostic restriction analysis of phAE8717W853G. Positive recombineering pools were used to transform XL-Blue cells and individual recovered phasmids used as substrates in a second round of MAMA-PCR. Independently recovered clones were tested by amplification of a 939 bp fragment containing the mutant region of TM4 gene 17, followed by digestion with Pvu I. The T→G substitution generates a new Pvu I site that is absent in the original sequence so that digestion generates 409 bp and 528 bp fragments. Uncut (U) and cut (C) products of two positive (#1 and 2) and two negative (#3 and 4) clones are shown.