Figure 2.
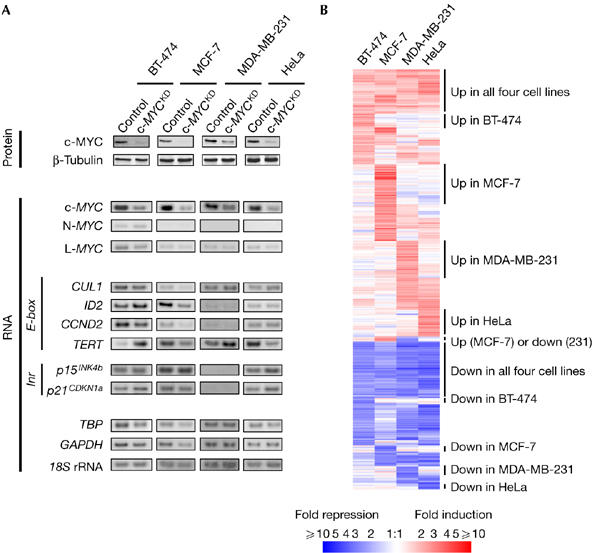
Transcriptional response to c-MYC depletion in human carcinoma cells. (A) c-MYC downregulation in the cells studied does not have a consistent effect on MYC targets expression. Proteins were collected 3 days after control and c-MYCKD (knockdown) short interfering RNA (siRNA) transfection and assayed for c-MYC downregulation. β-Tubulin analysis of the same extracts was used as a control for protein loading. Matching RNA samples were analyzed by quantitative radioactive reverse transcription-PCR for MYC family and MYC target (E-box- and Inr element-driven) genes expression: CUL1 (cullin 1), ID2 (inhibitor of DNA binding 2), CCND2 (cyclin D2), TERT (telomerase reverse transcriptase), P15INK4b (cyclin-dependent kinase 4 inhibitor B), P21CDKN1A (cyclin-dependent kinase inhibitor 1A). TBP (TATA box binding protein), GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and 18S rRNA were used for normalization. (B) Genome-wide patterns of transcriptional response to c-MYC depletion in human carcinoma cells. The cluster diagram depicts microarray data for genes that are regulated by siRNA-mediated c-MYC depletion. Mean fold-change values from independent replicates are given for 741 probe sets showing at least twofold regulation in one or more cell lines. White indicates unchanged expression, and red and blue shades indicate upregulation and downregulation, respectively, relative to control siRNA-transfected cells.
