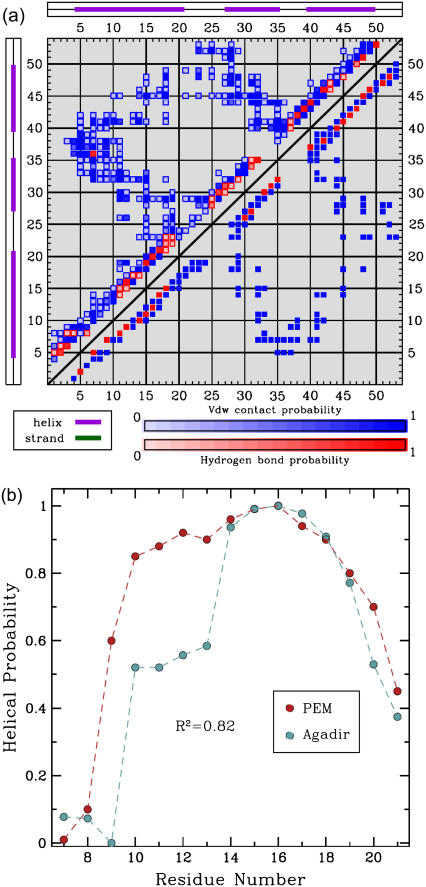
FIGURE 7.
(a) The contact map is drawn as a 53 × 53 square symmetric matrix (there are 53 aa in ALB8-GA). The formation of a contact between amino acids i, j is indicated with a blue square at position (i, j). The formation of a hydrogen bond between i, j is indicated with a red square at position (i, j). Shades of blue and red indicate different formation probabilities, with dark blue and dark red indicating a probability of 1, and lighter shades indicating lower probabilities. The top left half of the matrix shows the formation probabilities of contacts and hydrogen bonds in the PEM-generated ensemble. For reference, the bottom right of the matrix shows the contacts and hydrogen bonds in the representative NMR structure of ALB8-GA. The hydrogen bonds in the NMR structure indicate that amino acids Leu7-Lys11 are in helical configurations. The PEM-generated map shows that there are either missing or less probable hydrogen bonds in this region, indicating that Leu7-Lys11 visit unfolded configurations in the PEM-generated ensemble. (b) The probabilities for amino acids Leu7-Ala21 to be part of α1 are shown in red. These probabilities are measured over the ensemble conformations obtained by PEM. The secondary structure assignment for each conformation of the ensemble is computed with the STRIDE program (43) in the Tcl/TK environment of VMD (48). The normalized helicity scores predicted for each amino acid by Agadir (44) are shown in blue.