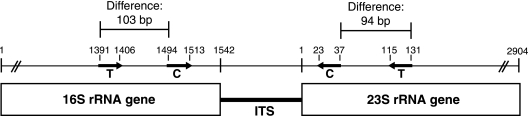
FIG. 1.
The positions of the primers used in this study are shown on the prokaryotic rRNA operon. Bold lines indicate primer annealing sites using E. coli numbering. T, 1406f/23Sr primers utilized by Fisher and Triplett (6); C, ITSF/ITSReub primers introduced by Cardinale et al. (3); ITS, 16S-23S rRNA operon internal transcribed spacer. Length heterogeneity in the ITS sequence is used to distinguish microbial populations in ARISA. The 1406f/23Sr primer set amplifies an additional 197 bases compared to that amplified by the ITS/ITSReub primer set (based upon standard E. coli numbering). This value was added to ITSF/ITSReub data to generate the cITSF/ITSReub data set and allowed direct comparison of data produced from the two primer sets.