FIG. 2.
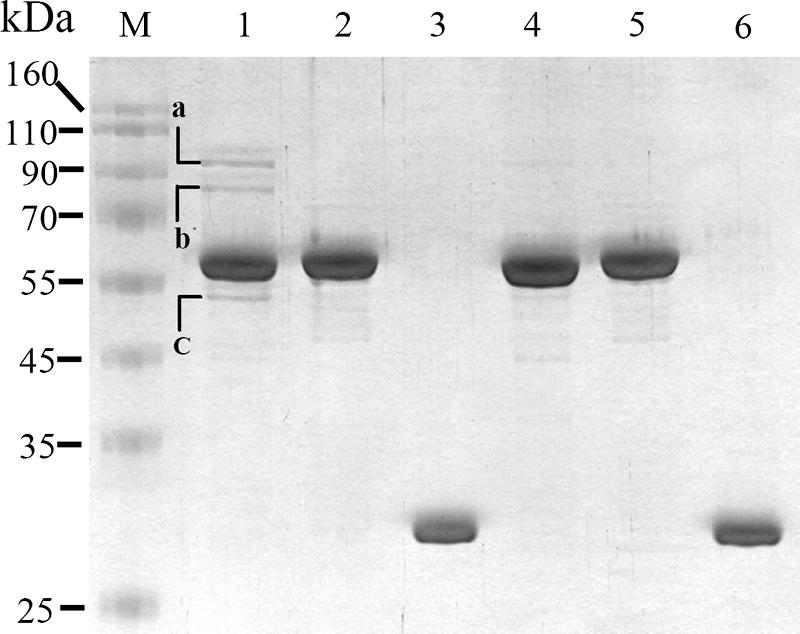
Use of protein pull-down assay for identification of S. marcescens proteins interacting with pirinSm. Glutathione-Sepharose 4B beads were used to capture protein complexes comprising GST-pirinSm fusion protein and the potential pirinSm interactors in CH-1. After elution, SDS-PAGE analysis was performed for protein separation. The cell lysate of E. coli DH5α containing either pSC10 (GST-pirinSm fusion protein was oversynthesized) (lane 1), pSC11 (GST-NlpBSm [28] fusion protein was oversynthesized) (lane 2), or pGEX (GST Tag only) (lane 3) was mixed with CH-1 lysate at a volume ratio of 1:1. Lane 4, GST-pirinSm fusion protein; lane 5, GST-NlpBSm fusion protein; lane 6, GST Tag protein; lane M, protein markers. Identification of the separated proteins was achieved by amino acid sequence analysis using electrospray ionization-MS/MS and comparison of the partial amino acid sequences with nonredundant protein databases in the SEQUEST Browser (http://fields.scripps.edu/sequest/). The three proteins with highest identity scores were E. coli PDH subunit E1 (band a, 99 kDa), E. coli PDH subunit E2 (band b, 76 kDa), and E. coli succinate dehydrogenase subunit E2 (band c, 50 kDa).
