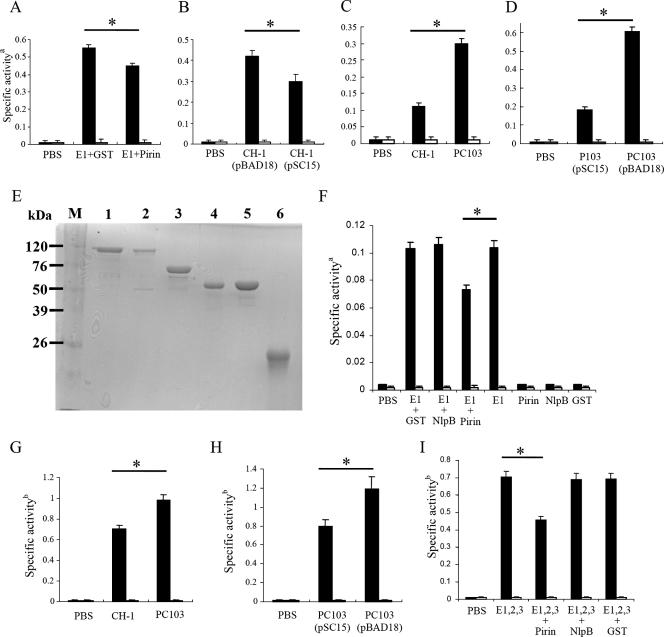
FIG. 4.
PirinSm inhibits PDH E1 and PDH enzyme complex activities. (A) Effect of pirinSm on PDH E1Sm specific activity. E. coli BL21(DE3) was used as the host for protein synthesis. (B) Effect of pirinSm gene overexpression (0.02% arabinose induction) from pSC15 on PDH E1Sm specific activity in S. marcescens CH-1. (C) Effect of pirinSm gene deletion on PDH E1Sm specific activity in S. marcescens CH-1. (D) Effect of pirinSm gene overexpression (0.02% arabinose induction) from pSC15 on PDH E1Sm specific activity in S. marcescens PC103. (E) Purification of S. marcescens PDH E1, E2, and E3 subunits in E. coli DH5α cells. Lane 1, GST-E1 subunit (pSC12); lane 2, GST-E2 subunit (pSC19); lane 3, GST-E3 subunit (pSC20); lane 4, GST-pirin (pSC10); lane 5, GST-NlpBSm (pSC11); 6, GST protein (pGEX vector). (F) Effect of purified pirinSm, NlpBSm, and GST (15 μg each) on specific activity of purified PDH E1 subunit (15 μg). (G) PDH enzyme complex activity in S. marcescens CH-1 and PC103. (H) Effect of pirinSm gene overexpression (0.02% arabinose induction) from pSC15 on specific PDH enzyme complex activity in S. marcescens PC103. (I) Effect of pirinSm on reconstituted PDH enzyme complex at an E1/E2/E3 molar ratio of 2:2:1. The DCPIP assay (12) was used to measure PDH E1 activity; the rate of NADH formation was measured spectrophotometrically at a wavelength of 340 nm (5) to measure PDH enzyme complex activity. Black and white bars indicate reactions with or without pyruvate added as the substrate. PBS (pH 7.4) was used as the negative control for all assays. CH-1, S. marcescens CH-1; PC103, a pirinSm gene insertion-deletion mutant strain derived from S. marcescens CH-1; pBAD18, pBAD18-Kan control vector; pSC15, pBAD18-Kan::pirinSm gene; NlpBSm, a membrane lipoprotein identified in S. marcescens CH-1 (28). a, change in OD600 per minute per milligram of protein. b, change in OD340 per minute per milligram of protein. Values are means and standard deviations from three independent experiments. *, P < 0.05.