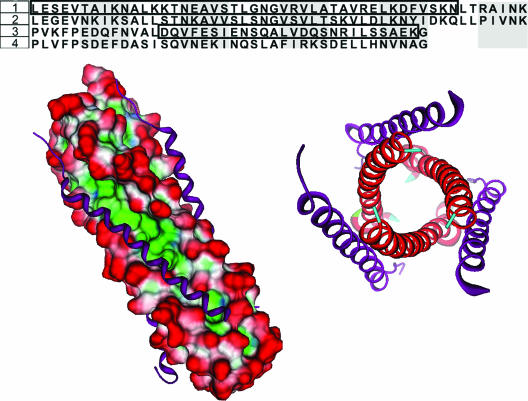
FIG. 2.
(Top) Sequence alignment of the HR regions of RSV and hMPV. RSV sequences were obtained from the crystal structure (PDB identification 1G2C). The HR-1 region shows an identity of 50% between the two viruses while the HR-2 region has 35% identity. 1, hMPV HR-1 (L130 to K179); 2, RSV HR-1; 3, hMPV HR-2 (P448 to G487); 4, RSV HR-2. LearnCoil-VMF-generated sequences for hMPV are boxed. (Bottom) The sequence of the RSV fusion core crystal was subjected to a strong local alignment with the hMPV sequence to determine appropriate homology sequences for the hMPV fusion core model. The alignment produced two hMPV peptides with 50% (HR-1) and 35% (HR-2) identity with the RSV peptides. Using the RSV structure as a template, the hMPV peptides were grafted onto the RSV Cα backbone and allowed to minimize. (Left) Shown is the surface of the HR-1 trimeric stalk (red, solvent exposed; blue, hydrophilic; green, hydrophobic) with HR-2 (purple ribbons) filling the hydrophobic grooves. (Right) Shown is an axial view of the hexameric core. The hexameric coiled-coil formation is the major structural feature in the postfusion conformation of the F protein.