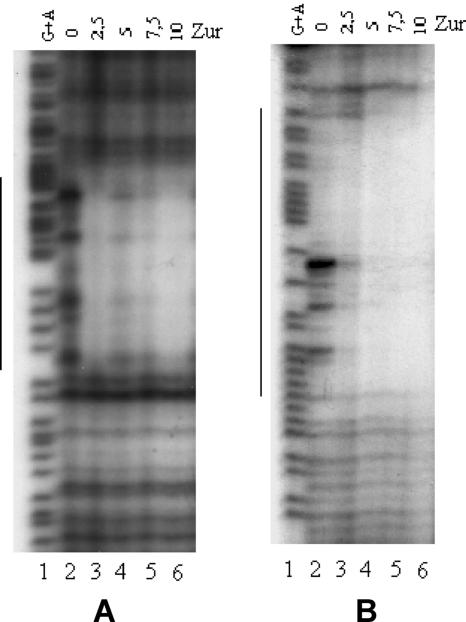
FIG. 4.
DNase I protection assays. (A) Footprint analysis of the Zur binding site upstream of Rv0280. The −156/+38 DNA region, digested with BamHI and ScaI restriction enzymes, was labeled at the −156 end, incubated in the presence of 0 to 10 μg of Zur for 20 min at room temperature, and finally digested with DNase I. Maxam-Gilbert A+G sequences of the same fragment were loaded in the first lane. (B) Footprint analysis of the Zur binding site upstream of rpmB2. The −181/+78 DNA region, digested with BamHI and XbaI restriction enzymes, was labeled at the +69 end, incubated in the presence of 0 to 10 μg of Zur protein for 20 min at room temperature, and finally digested with DNase I. Maxam-Gilbert A+G sequences of the same fragment were loaded in the first lane. Vertical bars on the sides of the gels indicate the protected regions.