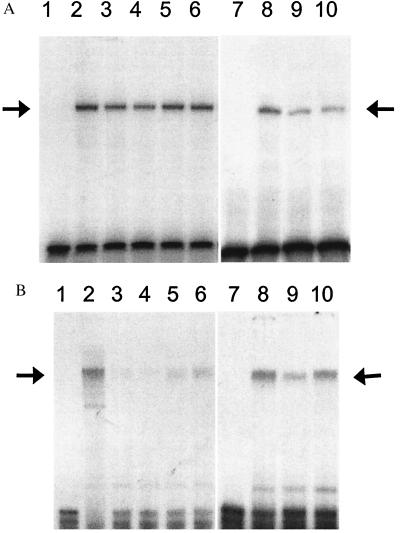
Figure 3.
Protein binding to the CT2 (A) and RIPE-3b1 (B) sites of the insulin gene promoter. Nuclear extracts from HIT-T15 cells chronically treated with NAC or AG were analyzed by EMSA, using a 32P-labeled human insulin promoter sequence −230 to −201 (A) and rat insulin gene II promoter sequence −126 to −101 (B). Lane 1, free probe. Lane 2, nuclear extract from HIT-T15 cells at p75. Lanes 3–6, nuclear extract from HIT-T15 cells at passage 107 chronically treated without (lane 3) or with 0.1 mM (lane 4), 0.5 mM (lane 5), and 1.0 mM (lane 6) of NAC. Lane 7, free probe. Lane 8, nuclear extract from HIT-T15 cells at p75. Lanes 9 and 10, nuclear extract from HIT-T15 cells at passage 104 chronically treated without (lane 9) or with 0.01 mM AG (lane 10). Arrows indicate the binding site of PDX-1/STF-1 (A) and RIPE-3b1 activator (B). The autoradiograms were quantitated by densitometry. The relative protein binding to the CT2 and RIPE-3b1 are normalized to the binding of control sample from HIT-T15 cells at passage 75. Relative CT2 binding: NAC 0, 0.61 ± 0.05; NAC 0.1, 0.58 ± 0.05; NAC 0.5, 0.79 ± 0.04*; NAC 1.0, 0.79 ± 0.05*; AG 0, 0.57; AG 0.01, 0.65. Relative RIPE-3b1 binding: NAC 0, 0.29 ± 0.04; NAC 0.1, 0.24 ± 0.61; NAC 0.5, 0.61 ± 0.10†; NAC 1.0, 0.79 ± 0.10†; AG 0, 0.56; AG 0.01, 0.85. Data are presented as mean ± SEM for four independent experiments at passage 105, 107, 108, and 109 in NAC study and are presented as mean of two independent experiments at passages 104 and 106 in AG study (*, P < 0.05 vs. NAC 0; †, P < 0.01 vs. NAC 0).