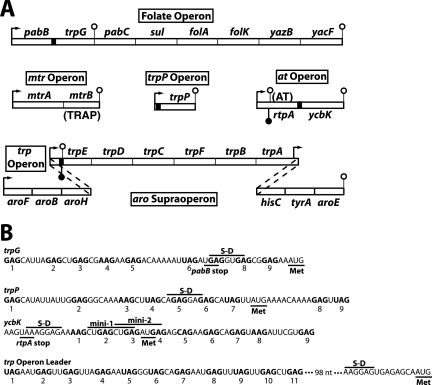
FIG. 1.
Organization of B. subtilis tryptophan metabolism genes and sequence comparison of the four known TRAP binding sites. (A) The six genes of the trp operon, which is part of the aromatic amino acid supraoperon, and trpG encode the tryptophan biosynthetic enzymes. trpP encodes a putative tryptophan transporter, while the ycbK gene product is similar to known efflux proteins. mtrA and the folate operon encode proteins involved in folic acid metabolism. mtrB encodes TRAP. TRAP regulates the expression of tryptophan metabolism genes by transcription attenuation (trpEDCFBA operon) and translation control (trpG, trpP, ycbK, and trpE) mechanisms. rtpA encodes anti-TRAP (AT), a protein that binds to and inactivates tryptophan-activated TRAP (25). Filled and open lollipop structures represent antiterminator and terminator structures, respectively. The four known TRAP binding sites are represented by black boxes, and bent arrows represent promoters. (B) The triplet repeats of the four known TRAP binding sites are shown in bold and are numbered. The S-D sequences and translation start codons (Met) are shown for trpG, trpP, ycbK, and trpE (first gene in the trp operon). There is a 98-nt gap in the sequence between the last triplet repeat in the trp operon leader and the trpE S-D sequence. The positions of the two dipeptide-encoding minigenes in the ycbK translation initiation region are marked. Expression of minigene 1 has a small inhibitory effect on translation of ycbK (28). The pabB and rtpA stop codons overlap the S-D sequences of trpG and ycbK, respectively.