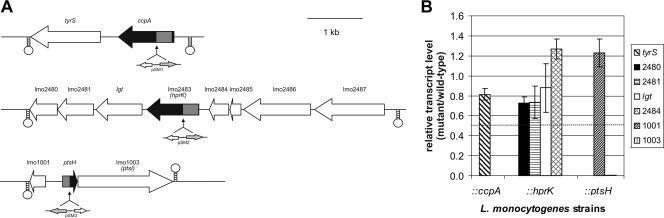
FIG. 2.
(A) Map of genetic loci disrupted during the present study with surrounding regions. All genes are drawn approximately to scale by using the L. monocytogenes EGD-e genome sequence data (http://genolist.pasteur.fr/ListiList/). The genes disrupted in L. monocytogenes EGD-e are indicated in black; adjacent ORFs are shown in white. The sites of pSM1-3 insertion into the corresponding genes are indicated by black arrows. Shaded areas within black arrows depict the regions of the genes cloned into pSM1-3. Stem-loop structures are used to illustrate putative terminator regions. Descriptions of the genes are as follows: tyrS, tyrosyl-tRNA synthetase; ccpA, catabolite control protein A; lmo2480, similar to acetyltransferase; lmo2481, similar to B. subtilis HPr-Ser-P phosphatase; lgt, highly similar to prolipoprotein diacylglyceryl transferase; lmo2483, HPr-Ser-P kinase/phosphatase; lmo2484, similar to B. subtilis YvlD protein; lmo2485, similar to B. subtilis YvlC protein; lmo2486, unknown; lmo2487, similar to B. subtilis YvlB protein; lmo1001, similar to B. subtilis protein YkvS; ptsH, PTS phosphocarrier protein HPr; lmo1003, phosphotransferase system enzyme I. (B) Transcriptional analysis with real-time RT-PCR to study the polar effect of pSM1-3 insertion on the transcription of genes located up- and downstream in the ccpA (tyrS), hprK (lmo2480, lmo2481, lgt, and lmo2484), and ptsH (lmo1001 and lmo1003) insertion mutants (indicated as ::ccpA, ::hprK, and ::ptsH, respectively) (see panel A). The strains were grown in BHI medium to an OD600 of 1.0, where the RNAs were prepared. The relative increases in the expression of the neighboring genes in the mutants compared to the wild-type strain (relative transcript level of mutant/wild-type) are depicted here. The relative expression levels of the genes studied was normalized to the housekeeping gene rpoB as described elsewhere (43, 60). The RT-PCR was performed with three independently isolated RNAs from the various strains in duplicate. The values represented here are means of the six obtained values, and the error bars indicate the standard deviations from the means. Relative expression levels of >1.8 or <0.55 (marked by dotted line) were considered to be differentially regulated based on microarray data (for details, see Material and Methods).