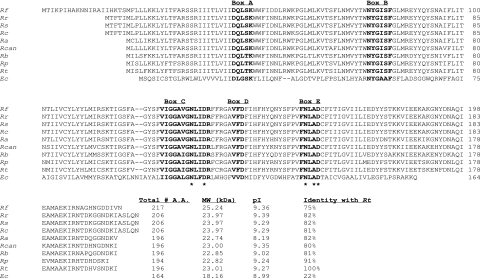
FIG. 1.
Alignment of deduced amino acid sequences of the lspA genes from R. typhi (Rt) (GenBank accession no. NC_006142), R. prowazekii (Rp) (GenBank accession no. AJ235271), R. bellii (Rb) (GenBank accession no. NZ_AARC01000001), R. canadensis (Rcan) (GenBank accession no. NZ_AAFF01000001), R. akari (Ra) (GenBank accession no. NZ_AAFE01000001), R. conorii (Rc) (GenBank accession no. NC_003103), R. sibirica (Rs) (GenBank accession no. AABW01000001), R. rickettsii (Rr) (GenBank accession no. NZ_AADJ01000001), R. felis (Rf) (GenBank accession no. NC_007109), and E. coli (Ec) (GenBank accession no. X00776). Molecular masses and isoelectric points were computed with MacVector 7.1.1 software. The conserved amino acid domains (boxes A through E) are shown in bold. The catalytic residues are marked by asterisks.