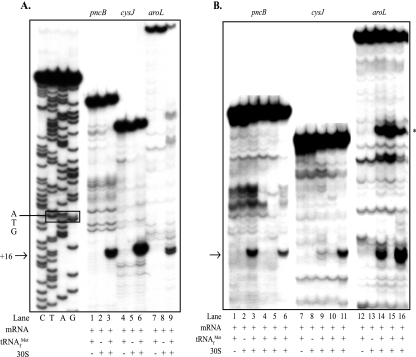
FIG. 2.
Effects of aroL, pncB, and cysJ downstream adenines on ribosome binding in vitro: toeprint assays with aroL-, pncB-, and cysJ-lacZ mRNAs and 30S subunits comparing ribosome binding to mRNAs with and without mutations altering the downstream adenine content (Table 1). The arrows indicate the position of the ternary complex-dependent toeprint signals at position 16 relative to the A residue (position 1) of the AUG start codons that result from bound 30S subunits, and the asterisk in panel B indicates the position of an uncharacterized ternary complex-dependent signal in the aroL upstream UTR. Bands observed in the absence of tRNA and ribosomes were artifacts likely due to structures formed within the mRNA. (A) Lanes G, A, T, and C, dideoxy DNA sequence ladder for the pncB template (the ATG initiation triplet is enclosed in a box); lanes 1 to 3, pncB-lacZ wild-type mRNA; lanes 4 to 6, cysJ-lacZ wild-type mRNA; lanes 7 to 9, aroL-lacZ wild-type mRNA. (B) Lanes 1 to 6, pncB-lacZ mRNAs made from the templates pApncB.WT (lanes 1 to 3), pApncB.DN1b (lane 4), pApncB.DN1 (lane 5), and pApncB.UP1 (lane 6); lanes 7 to 11, cysJ-lacZ mRNAs made from the templates pAZcysJ.WT (lanes 7 to 9), pAZcysJ.DN1 (lane 10), and pAZcysJ.UP1 (lane 11); lanes 12 to 16, aroL-lacZ mRNAs made from the templates pAaroL.WT (lanes 12 to 14), pAaroL.DN1 (lane 13), and pAaroL.UP1 (lane 14).