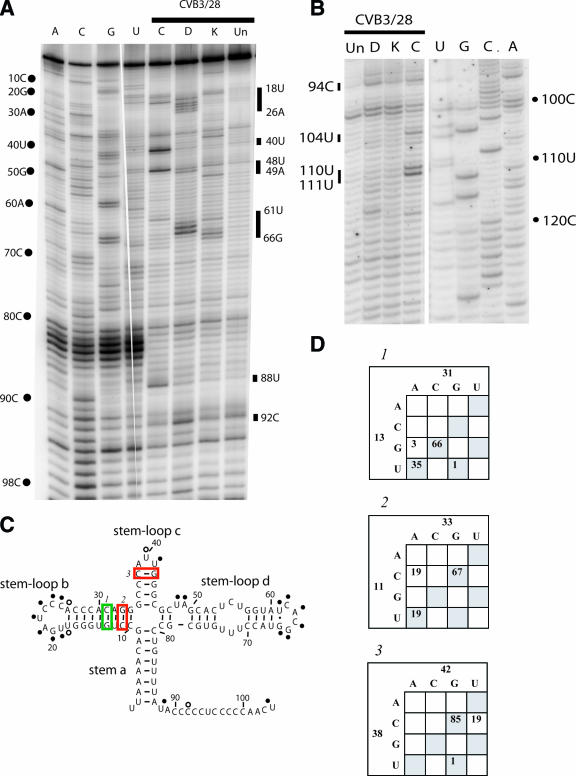
FIG. 2.
Chemical probing results for domain I (cloverleaf). A. A 12% sequencing gel showing primer extension analysis of modified and unmodified CVB3 RNA. Labels on the left indicate nucleotide positions according to the sequencing tracks (lanes A, C, G, and U), and labels on the right identify positions that are modified. Lane Un, unmodified; lane K, kethoxal; lane D, DMS; lane C, CMCT. B. A 12% sequencing gel to show detailed results for the connecting region between domain I and domain II. Labels on the right indicate nucleotide positions according to the sequencing tracks (lanes U, G, C, and A), and labels on the left identify positions that are modified. Lane Un, unmodified; lane K, kethoxal; lane D, DMS; lane C, CMCT. C. Predicted secondary structure map of domain I, showing modified positions. Filled circles identify strongly modified positions; open circles identify moderately modified positions. Red boxes indicate examples of pairs that are not supported by the comparative sequence analysis results shown in panel D. Green boxes indicate examples of pairs that are supported by phylogentic analysis in panel D. D. Analysis of representative paired positions, showing the number of occurrences of nucleotide identities among 105 enterovirus sequences.