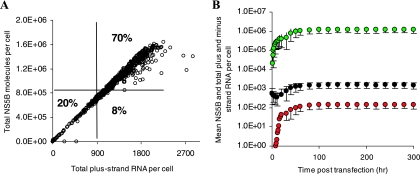
FIG. 3.
Sensitivity analyses for the model of subgenomic HCV RNA replication in Huh-7 cells. One thousand parameter sets within the parameter ranges given in Table 1 (column 3) were randomly chosen. (A) Within these parameter ranges we found that: (i) ∼20% of parameter sets led to a total plus-strand RNA steady state of <900 copies/cell or HCV RNA elimination, (ii) ∼8% led to a total plus-strand RNA steady state of >900 copies/cell but NS5B molecules of <8 × 105, (iii) ∼70% (700) of parameter sets generated steady states of between 900 and 5,000 total plus-strand RNAs and 8 × 105 to 4 × 106 NS5B molecules, and (iv) ∼2% led to a total plus-strand RNA steady state of >5,000 copies/cell (not shown). Among these 700 parameter sets only 85% (600 sets) attained an RNA steady-state in 48 h posttransfection (data not shown). (B) Using the interquartile ranges for each parameter in those 600 sets (Table 1, column 4), we found that more than 99% of 1,000 simulations using randomly chosen parameters within these ranges of the parameter led to steady states consistent with the characteristics given in Materials and Methods. Mean values of total plus-strand RNA, minus-strand RNA, and NS5B molecules are shown in black, red, and green filled circles, respectively. Vertical lines represent three standard deviations.