Abstract
Although the Janus kinase-signal transducer and activator of transcription (JAK-STAT) signaling pathway is part of the antiviral response in arthropods such as Drosophila, here we show that white spot syndrome virus (WSSV) uses a shrimp STAT as a transcription factor to enhance viral gene expression in host cells. In a series of deletion and mutation assays using the WSSV immediate-early gene ie1 promoter, which is active in shrimp cells and also in insect Sf9 cells, an element containing a STAT binding motif was shown to be important for the overall level of WSSV ie1 promoter activity. In the Sf9 insect cell line, a specific protein-DNA complex was detected by using electrophoresis mobility shift assays (EMSA) with the 32P-labeled STAT binding motif of the WSSV ie1 promoter as the probe. When recombinant Penaeus monodon STAT (rPmSTAT) was overexpressed in Sf9 cells, EMSA with specific antibodies confirmed that the STAT was responsible for the formation of the specific protein-DNA complex. Another EMSA showed that in WSSV-infected P. monodon, levels of activated PmSTAT were higher than in WSSV-free P. monodon. A transactivation assay of the WSSV ie1 promoter demonstrated that increasing the level of rPmSTAT led to dose-dependent increases in ie1 promoter activity. These results show that STAT directly transactivates WSSV ie1 gene expression and contributes to its high promoter activity. We conclude that WSSV successfully annexes a putative shrimp defense mechanism, which it uses to enhance the expression of viral immediate-early genes.
White spot syndrome virus (WSSV) is the type species of the genus Whispovirus, family Nimaviridae (40). WSSV is a large DNA virus with a virion that consists of a nucleocapsid, tegument, and envelope and includes at least 39 structural proteins (22, 38, 39, 43). WSSV is extremely virulent (17, 28, 29), has a wide host range (12, 26), and targets various tissues (25, 26, 42). The rapid onset and lethality of white spot disease are remarkable (8). Replication of the virus is easily triggered by physiological or environmental stress, but partly because no continuous shrimp cell line is currently available, the molecular mechanisms that control WSSV gene transcription are still largely unknown.
Recently, three WSSV immediate-early (IE) genes (ie1, ie2, and ie3) were identified by microarray and reverse transcription-PCR analysis in cycloheximide-treated WSSV-infected shrimp (24). WSSV ie1 exhibits very strong promoter activity and is also unusual in that it is highly expressed throughout the infection cycle. WSSV ie1 promoter activity is also very strong in Sf9 insect cells, which are used for studying WSSV genes at the cellular level even though they are not permissive to WSSV (16, 24, 27). Although all of the functions of these WSSV IE genes are not yet known, IE1 has a Cys2/His2-type zinc finger DNA binding motif (24) that is functionally active (unpublished data), and in general, viral immediate-early genes are critically important in the virus infection cycle. The expression of viral IE genes is also known to depend on the host cell's transcription and translation machineries (6).
On the host side, members of the signal transducer and activator of transcription (STAT) family play a vital role in the innate immunity of both vertebrates and invertebrates (1, 2, 5, 20). STAT has recently been shown to be an important part of Drosophila's defense response to viral infection (11). The Janus kinase (JAK)-STAT pathway is also activated in response to bacterial infection in mosquitoes (5, 23), and it responds to stresses such as oxidative stress in rat liver (35). In the present study, by functionally mapping a series of deletions of the ie1 promoter, we found that a fragment with the consensus STAT binding motif was critical for the promoter function. To further functionally characterize this motif, we used a protein-DNA binding assay with site-directed mutagenesis, electrophoretic mobility shift assays (EMSA), and transactivation analysis with a recombinant shrimp STAT. To our knowledge, this is the first report to investigate the transcriptional control of WSSV genes.
MATERIALS AND METHODS
Virus.
A WSSV Taiwan isolate (WSSV T-1; GenBank accession no. AF440570) and the baculovirus Autographa californica multiple nucleopolyhedrosis virus (AcMNPV) (GenBank accession no. NC_001623) were used as templates for amplification of their respective ie1 gene promoters.
Transient transfections and dual luciferase reporter assay.
For the promoter activity assays in this study, we used Sf9 insect cells and a dual luciferase reporter assay. For DNA transfection, the Sf9 insect cells were seeded onto a 24-well plate (3 × 105 cells/well) and grown in Sf-900 II SFM serum-free medium (Invitrogen) overnight at 27°C. Plasmids containing the firefly luciferase gene (including the pGL3-Basic firefly luciferase reporter vector [Promega], which was used as a negative control) were transfected into the Sf9 cells by using the Cellfectin reagent (Invitrogen) (1 μg of plasmid DNA per well), and this was followed by cotransfection with 100 ng of the Renilla luciferase plasmid phRL/AcMNPVie1. This plasmid, which contained the Renilla luciferase reporter gene, was constructed by cloning the AcMNPV ie1 promoter into the phRL-null vector (Promega) by using primer pair AcMNPV-ie1-F1/AcMNPV-ie1-R1 (Table 1), and it was used to monitor and normalize transfection efficiency. Cells were collected at either 24 or 48 h after transfection, and cell lysates were prepared according to the Promega instruction manual for the dual luciferase assay system. Luciferase activity was measured with a luminometer (Labsystems). Firefly luciferase activity values were then normalized against the activities of the Renilla luciferase to correct for transfection efficiency, and data were expressed as relative luciferase activity. Independent triplicate experiments were performed for each plasmid, and the mean and standard deviation (SD) were calculated.
TABLE 1.
Primers used for generating deletion mutants and other clones for WSSV ie1 promoter activity assays
| Plasmida | Primersb |
|---|---|
| p(−2011/+52) | Fwd1 (CGGGTACCGATGATGGTGATGTTTCTAGG) |
| Rev1 (CCGCTCGAGCTTGAGTGGAGAGAGAGAGC) | |
| p(−945/+52) | Fwd2 (CGGGTACCGAGATCCTAGAAAGAGGAGTG)/Rev1 |
| p(−703/+52) | Fwd3 (CGGGTACCGGACAGTAGAGGGTTATACG)/Rev1 |
| p(−450/+52) | Fwd4 (CGGGTACCGTGGCTAATGGAGAATTGTCGT)/Rev1 |
| p(−268/+52) | Fwd5 (CGGGTACCGGTGTTAAAGAAGCAGTTGTG)/Rev1 |
| p(−94/+52) | Fwd6 (CGGGTACCCCTTGTTACTCATTTATTCCTAG)/Rev1 |
| p(−71/+52) | Fwd7 (CGGGTACCAAATGGTGTAATCGCTGTTG)/Rev1 |
| p(+5/+52) | Fwd8 (CGGGTACCCCCGTGTTAGCTCCTCGAT)/Rev1 |
| phRL/AcMNPVie1 | AcMNPV-ie1-F1 (TCGATGTCTTTGTGATGCGC) |
| AcMNPV-ie1-R1 (AACTTGCAACTGAAACAATATC) | |
| p(−945/+52)23mer-del | Fwd2/Rev2 (CGGGTACCAAATTCCTTAACATGATTCA) |
| p(−268/+52)23-mer-del | Fwd5/Rev2 |
| pN(−72/−94) | CCCTTGTTACTCATTTATTCCAGGGTAC |
| CCTAGGAATAAATGAGTAACAAGGGGTAC | |
| pC(−94/−72) | GATCCCCTTGTTACTCATTTATTCCTAGG |
| GATCCCTAGGAATAAATGAGTAACAAGGG | |
| pC(−72/−94) | GATCCCCTTGTTACTCATTTATTCCTAGG |
| GATCCCTAGGAATAAATGAGTAACAAGGG | |
| p(−94/+52)STAT4mer-mut | Fwd6-STAT4mer-mut (CGGGTACCCCTTGTTACTCATTTAGGCCTAGCCA)/Rev1 |
| p(−94/+52)STAT2mer-mut | Fwd6-STAT2mer-mut (CGGGTACCCCTTGTTACTCATTTATTCCTAGCCA)/Rev1 |
WSSV ie1 nucleotide positions are from reference 24.
Sequences are 5′ to 3′. Added restriction enzyme cutting sites are underlined.
Functional deletion assay of WSSV ie1 promoter activity in Sf9 insect cells.
To analyze the WSSV ie1 basal promoter and regulatory regions, progressive 5′ deletions were made on the promoter region. Using a universal reverse primer (Rev1) and eight different forward primers (Table 1), PCR was used to generate fragments that started at different positions and ended at nucleotide (nt) +52 relative to the transcription start site (+1). These DNA fragments, which contained the WSSV ie1 promoter region and had KpnI and XhoI restriction enzyme cutting sites at either end, were then cloned into the pGL3-Basic firefly luciferase reporter vector to produce the constructions shown in Fig. 1. Dual luciferase reporter assays were then conducted as described above, and luciferase activities were measured using a dual luciferase assay system.
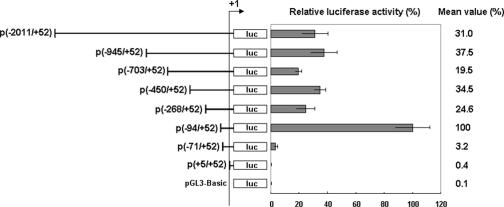
FIG. 1.
Functional mapping of deletions of the WSSV ie1 promoter. Relative luciferase activity has been normalized to the activity of the p(−94/+52) vector, which was arbitrarily set to 100%. Data represent the means from triplicate experiments. Error bars show the SDs. The plasmid numbers in parentheses specify the beginning and end positions of the promoter fragments, and the arrow labeled +1 marks the transcription start site. The difference between the p(−94/+52) and p(−71/+52) plasmids is a 23-mer fragment that consists of an imperfect inverted repeat, −94CCTTGTTACTCATTTATTCCTAG−72.
Deletion of a 23-mer fragment on the WSSV ie1 promoter.
From the results of the progressive 5′-deletion assay, we identified a 23-mer fragment from nt −94 to −72 that had a strong effect on WSSV ie1 promoter activity. To further elucidate the importance of this sequence, two more plasmids were constructed as follows: DNA fragments −945/−95 and −268/−95 were amplified by PCR using the primer pairs Fwd2/Rev2 and Fwd5/Rev2 and then subcloned into the KpnI site of the p(−71/+52) plasmid to produce two mutated deletion plasmids, p(−945/+52)23mer-del and p(−268/+52)23mer-del (Table 1). The activities of these two deletion mutants were then measured by dual luciferase activity assay.
Testing the enhancer activity of the WSSV ie1 promoter 23-mer fragment.
To determine whether the 23-mer fragment also acts as an enhancer, three additional plasmids were constructed as follows. Sense and antisense oligonucleotides of the 23-mer fragment were synthesized and then modified so that one end included either part of the KpnI recognition site sequence or a BamHI site (Table 1). The oligonucleotide strands were denatured by heating in boiling water for 10 min and then annealed by slowly cooling to room temperature. The annealed DNA fragments were treated with T4 polynucleotide kinase (Promega) and ligated in either the sense or antisense orientation with plasmid p(−71/+52) after it had been digested with either KpnI or BamHI. This resulted in an antisense 23-mer fragment being located in front of the N-terminal basal promoter region (KpnI digestion) for plasmid pN(−72/−94) or else in a sense or antisense fragment behind the C-terminal poly(A) addition site (BamHI digestion) for plasmids pC(−94/−72) and pC(−72/−94), respectively. For the complement of pN(−72/−94) in this assay, the p(−94/+52) plasmid (described above) was redesignated pN(−94/−72), as these two plasmids are exactly equivalent. Dual luciferase activity assays were then conducted as described above.
Computer analysis of potential transcription factor binding sites in the 23-mer fragment.
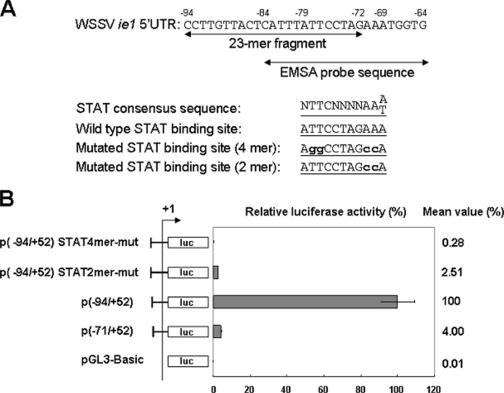
Transcription Element Search software (TESS) (http://www.cbil.upenn.edu/tess) was used to search for transcriptional factor binding sites within and near to the 23-mer fragment. The TESS program, which exploits the TRANSFAC database, identified a putative STAT motif from nt −69 to −79 upstream of the transcription start site.
Dual luciferase activity assay of the wild-type versus mutated putative STAT binding site between nucleotides −79 and −69 on the ie1 promoter.
To further confirm that the putative STAT binding motif contributes to the high expression level of WSSV ie1, a mutation assay was performed. Site-directed mutagenesis was used to change the gamma interferon-activated sequence element of the consensus STAT binding sequence (5′-NTTCNNNNAAA/T-3′) (33) from the wild type 5′-ATTCCTAGAAA-3′ to the 4-mer mutant 5′-AGGCCTAGCCA-3′ and the 2-mer mutant 5′-AGGCCTAGCCA-3′ (the mutated nucleotides are shown in boldface). The plasmids p(−94/+52)STAT4mer-mut and p(−94/+52)STAT2mer-mut, respectively, were constructed from these mutated sequences and then used in dual luciferase activity assays as described above. The sequences of the Fwd6-STAT4mer-mut, Fwd6-STAT2mer-mut, and Rev1 primers used for the construction of the mutated clones are listed in Table 1.
EMSA for binding of STAT to the putative ie1 STAT binding motif.
For the EMSA experiments, first a heat shock-inducible plasmid was constructed to overexpress recombinant Penaeus monodon STAT (rPmSTAT) in transfected Sf9 insect cells. Construction was as follows. The heat-inducible promoter of Drosophila melanogaster hsp70 (nt −525 to +236 according to Torok and Karch ([37]) (GenBank accession no. J01103) was PCR amplified from D. melanogaster genomic DNA by using the primers 5′-CCTCATGAGTTGACAACAACAGTCTTGACAACCT-3′ and 5′-GGAAGCTTCCTCGGTAACGACTTGTTGAAAGT-3′, where the underlined nucleotides are BspHI and HindIII recognition sequences, respectively. The PCR-amplified hsp70 promoter DNA fragments were then digested with the BspHI and HindIII and ligated with pIZ/V5-His plasmid (Invitrogen) after it had been digested with BspHI and HindIII to release the OpIE2 promoter upstream multiple cloning sites. The resulting plasmid was designated pDhsp/V5-His. To insert STAT downstream of the heat shock promoter, the full-length P. monodon STAT gene was cloned by PCR from P. monodon cDNA by using the primers 5′-GGAAGCTTCACAATGTCGTTGTGGAACAG-3′ and 5′-GGAAGCTTTTATGAAAAGTCTGAGAGG-3′, where the underlined sequences indicate HindIII recognition sites. (The P. monodon STAT gene was cloned and sequenced in a previous study and had been submitted to the GenBank database with accession no. AY327491.) The PmSTAT cDNA was then cloned into the HindIII-digested pDhsp/V5-His plasmid to produce pDhsp/PmSTAT/V5-His. The structure of this plasmid was confirmed by DNA sequencing.
These plasmids were then used to transfect Sf9 cells and prepare nuclear extracts as follows. Sf9 cells were transfected for 24 h with either pDhsp/V5-His (control plasmid with no PmSTAT insertion) or pDhsp/PmSTAT/V5-His. The cells were then heat shocked (42°C water bath for 30 min), and after a further 2, 4, or 6 h, nuclear extracts of the cells were harvested by the modified Dignam method as described previously (34). Protein levels were then quantified using a Bio-Rad protein assay kit (Bio-Rad). For the EMSA and other assays, nuclear extracts were also prepared from uninfected and WSSV-infected (72 h postinfection [hpi]) P. monodon gill tissue by using the method described by Lahiri and Ge (21). The Bio-Rad protein assay kit was used to quantify protein levels in these extracts also.
For the EMSA to test the ability of PmSTAT to bind with the putative ie1 STAT binding sequence, the Sf9 and shrimp gill tissue nuclear extracts were separately mixed with EMSA reaction buffer [4% Ficoll, 12 mM HEPES (pH 7.9), 4 mM Tris-HCl (pH 7.9), 0.1 mM EDTA, 5 μg of poly(dI-dC) and 1 mM dithiothreitol] to a total volume of 15 μl and incubated for 10 min at room temperature. A 32P-labeled (30,000 cpm) double-stranded ie1 promoter probe (5′-−84CATTTATTCCTAGAAATGGTG−64-3′; the gamma interferon-activated sequence element is underlined) was added, and the mixture was incubated at room temperature for 20 min. For the competition experiments, nuclear extracts were preincubated for 10 min with a 10× or 40× molar excess of the following unlabeled double-stranded competitor oligonucleotides: wild-type STAT binding motif (5′-TTATTCCTAGAAATG-3′), the 4-mer mutated STAT binding motif (5′-TTAGGCCTAGCCATG-3′; the mutated nucleotides are shown in boldface), the SfSTAT binding motif (5′-TGTTCTGAGAAA-3′) (SfSTAT is our designation for SPI-GLE 1, which has previously been shown to have a STAT-like binding activity in Sf9 cells [33]), the AP-1 binding motif (5′-CGCTTGATGAGTCAGCCGGAA-3′) and the Oct-1 binding motif (5′-TGTCGAATGCAAATCACTAGAA-3′). After preincubation, the labeled probe was added and the mixtures were incubated as described above (room temperature; 20 min). The reaction mixtures were then separated by 6% polyacrylamide gel electrophoresis (PAGE), after which the gels were dried and exposed to Kodak Biomax MS film.
Supershift EMSA was used to confirm that the EMSA complexes were specifically formed between PmSTAT and the ie1 STAT binding motif. For the supershift assays, nuclear extracts prepared as described above were preincubated for 1 h at room temperature in the presence of either rabbit anti-V5 polyclonal antibody (Sigma) or mouse anti-glutathione S-transferase (anti-GST) monoclonal antibody (Sigma) (for Sf9 cells) or else with anti-rPmSTAT antiserum (for P. monodon gill tissues). The labeled probe was then added to each mixture, and the mixtures were incubated as described above. Separation was by 4% PAGE, and this was followed by drying and exposure to Kodak Biomax MS film.
The anti-rPmSTAT antiserum used in the supershift assay was prepared as follows. A DNA fragment representing the coding region of PmSTAT was PCR amplified by using the primers 5′-CGGGATCCATGGGTTTTGTGGGTCGACGCCA-3′ and 5′-CCCAAGCTTTGAAAAGTCTGAGAGGACATTTG-3′. After amplification, the PCR products were digested with restriction enzymes, cloned into pET-28b(+) (Novagen), and transformed into BL21 Codon Plus Escherichia coli cells (Stratagene). For protein expression and purification, the cells were grown overnight at 37°C in Luria-Bertani medium supplemented with 50 μg/ml of kanamycin and 34 μg/ml of chloramphenicol. The cells were inoculated into new medium at a ratio of 1:300 and grown at 37°C for 1.5 to 2 h. Protein expression was induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and incubation was continued for another 1.5 to 3 h. The induced bacteria were spun down at 4°C, suspended in ice-cold phosphate-buffered saline (PBS) containing 10% glycerol and a protease inhibitor cocktail tablet (Roche), and sonicated for 30 s on ice. The insoluble debris was collected by centrifugation, suspended in phosphate-buffered saline containing 1.5% sodium lauryl sarcosine, and solubilized by shaking at 4°C for 2 h. After being clarified by centrifugation, the supernatant was mixed with Ni-nitrilotriacetic acid-agarose beads (QIAGEN) on a rotating wheel at 4°C for 16 h. The beads were then washed several times with ice-cold wash buffer (1 M NaCl, 10 mM Tris-HCl, pH 7.5) to remove unbound material. The fusion proteins were eluted directly from the beads with sodium dodecyl sulfate (SDS) sample buffer and then subjected to SDS-PAGE analysis. The protein bands containing the fusion proteins were sliced from the gel, minced, mixed with Freund's adjuvant, and used for antibody production.
Phosphorylation status of PmSTAT.
To determine the phosphorylation (activation) status of PmSTAT, we used immunoprecipitation and Western blot analysis with either the anti-rPmSTAT antiserum (prepared as described above) or antiphosphotyrosine antibody (Upstate). For these assays, nuclear extracts were prepared as described above, and equal amounts of nuclear proteins from all of the nuclear extracts were separately dissolved in radioimmunoprecipitation assay (RIPA) buffer (0.5% [vol/vol] NP-40, 0.5% [wt/vol] sodium deoxycholate, 0.5% [wt/vol] SDS, 150 mM NaCl, 10 mM Tris-HCl [pH 7.4], 5 mM EDTA, 0.1 mM Na3VO4, and a protease inhibitor cocktail tablet). These RIPA buffer-dissolved preparations were incubated with either 1 μl of anti-V5 antibody (for Sf9 samples) or 1 μl of preimmune rabbit serum or anti-rPmSTAT antiserum (for P. monodon gill preparations) and then precipitated with 50 μl of protein A-agarose (Invitrogen). After reaction at 4°C overnight, the agarose beads were washed extensively with RIPA buffer and the bound proteins were fractionated by 7.5% SDS-PAGE, transferred to a polyvinylidene difluoride membrane (MSI), incubated with either anti-rPmSTAT antibody or antiphosphotyrosine antibody, and then detected with a secondary peroxidase-conjugated antibody. Detected proteins were visualized using an ECL (Perkin-Elmer) detection system.
Laser scanning microscopy for cell localization assay of pDhsp/PmSTAT/V5-His transfected Sf9 cells.
Monolayers of Sf9 cells were transfected with pDhsp/PmSTAT/V5-His for 24 h and then heat shocked (42°C water bath for 30 min). At 6 h after heat induction, the monolayers were washed twice with PBS, and the cells were fixed in paraformaldehyde (4% in PBS) for 10 min at 4°C, treated with 0.1% Triton X-100 in 4% paraformaldehyde-PBS solution (3 min at 4°C), and then washed thoroughly two times with PBS. After blocking in bovine serum albumin and normal goat serum (diluted to 5% and 2% in PBS, respectively) for 16 h at 4°C, the cells were treated with a 3,000× PBS-diluted polyclonal rabbit anti-V5 antibody (3 h at room temperature). The cells were then washed with 0.2% Tween 20 in PBS (three washes of 10 min each) and reacted with 1,000× PBS-diluted carboxymethylindocyanine (Cy3) dye-conjugated goat anti-rabbit immunoglobulin G antibody (Sigma) for 2 h at room temperature. Counterstaining of the nucleus was performed with 4′,6′-diamidino-2-phenylindole dihydrochloride (DAPI) (Vector Laboratories). After washing three times with 0.2% Tween 20 in PBS (10 min each time), the cover glasses were wet mounted with 60% glycerol. During all of the above-described reactions, the monolayers were kept in darkness. Fluorescent signals were examined using a confocal laser scanning microscope (Leica TCS SP2).
PmSTAT transactivation activity assay.
For the transactivation experiments, Sf9 cells were cotransfected using 500 ng of the reporter plasmid containing the WSSV ie1 promoter [p(−94/+52)], 100 ng of the phRL/AcMNPVie1 internal control plasmid, and different quantities (0, 20, 100, 200, 400, and 500 ng) of pDhsp/PmSTAT/V5-His heat-inducible expression plasmid. The total amount of DNA was kept constant by adding an appropriate amount of pDhsp/V5-His control plasmid. For comparison, a parallel set of experiments were run using 500 ng of the p(−94/+52)STAT4mer-mut plasmid instead of the ie1 p(−94/+52) promoter plasmid. At 48 h posttransfection, the Sf9 cells were heat shocked, and 6 h later the cells were harvested and lysed. Luciferase activity in the cell extracts was measured as described above.
RESULTS
A 23-mer fragment is critical for the WSSV ie1 promoter activity.
The functional deletion mapping results (Fig. 1) show a large drop (∼31×) in luciferase activity from a nominal 100% for p(−94/+52) down to a basal level (3.2%) for p(−71/+52). The difference between these two deletions is a critical 23-nt fragment that is an imperfect inverted repeat, 5′-−94CCTTGTTACTCATTTATTCCTAG−72-3′. Figure 1 also shows that in the negative control plasmid p(+5/+52), deletion of the TATA box and transcription start site resulted in negligible activity levels. We also note in passing that a TESS database computer analysis of the region between nt −94 and −268 found binding sequences for the potential regulatory proteins Ftz, Pax-5, RelA, and NF-AT, which may account for the substantial drop in activity for p(−268/+52) and for the other constructs with longer promoter regions.
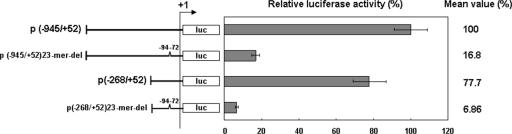
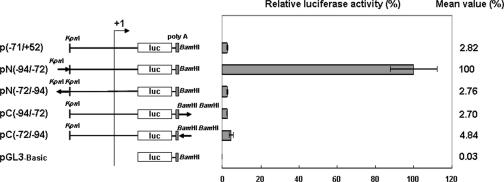
The importance of the 23-mer fragment was confirmed by another deletion assay (Fig. 2). Here, the luciferase activities of the two 23-mer deletion plasmids, p(−945/+52)23mer-del and p(−268/+52)23mer-del, were, respectively, 6 and 11 times lower than those of the corresponding plasmids that contained the 23-mer fragment, p(−945/+52) and p(−268/+52). No enhancement of activities was observed in the pN(−72/−94), pC(−94/−72), and pC(−72/−94) plasmids (Fig. 3). These plasmids produced luciferase activities that were similar to the basal levels produced by p(−71/+52), which shows that the 23-bp region upstream of the basal promoter does not work as an enhancer.
FIG. 2.
Effect of deleting the 23-mer fragment from the WSSV ie1 promoter region. The two deletion constructs, p(−945/+52)23mer-del and p(−268/+52)23-mer-del, are both missing the −94/−72 region of the WSSV ie1 promoter. Data show the means of three repetitions, and error bars show the SDs.
FIG. 3.
Testing the enhancer activity of the 23-mer fragment. The solid arrows show the locations and orientations (sense or antisense) of the 23-mer fragment in two complementary plasmid pairs. In this figure, the (−94/+52) plasmid has been redesignated pN(−94/−72). Results are shown as means ± SDs from three independent experiments.
Computer searches for transcriptional factor binding motifs did not initially find any matches for the 23-mer fragment, but when neighboring sequences were included, TESS search results predicted a transcriptional factor binding site for STAT (Fig. 4A). We hypothesized that this putative STAT binding site might be critically important for the high levels of WSSV ie1 promoter activity, and the following assays were designed to test this hypothesis.
FIG. 4.
Site-directed mutagenesis of the STAT binding motif in the WSSV ie1 promoter region. (A) Locations and sequences of the 23-mer fragment and EMSA probe and the STAT consensus sequences. Sequences of the consensus, wild-type, and mutated STAT binding sites are shown. The boldface lowercase letters indicate the mutated nucleotides. (B) Relative luciferase activities of WSSV ie1 p(−94/+52) promoter constructs with wild-type or mutated STAT binding sites. Data represent the means ± SDs from three independent experiments.
The STAT binding site (ATTCCTAGAAA) contributes to WSSV ie1 promoter activity.
A site-directed mutation assay was performed to confirm the relationship between the STAT binding motif and the high expression level of WSSV ie1. Figure 4B shows marked decreases in the luciferase activities of the constructs in which the putative WSSV ie1 promoter STAT binding site was mutated (Fig. 4A). Compared to the p(−94/+52) plasmid, activity levels of the two mutants fell by factors of approximately 357× and 40×, which were, respectively, about 14× and 2× lower even than the p(−71/+52) basal promoter driver activity in the Sf9 cells. These data suggest that the STAT binding site that overlaps the 23-mer fragment functions as a cis-acting element of the WSSV ie1 promoter.
EMSA detects a specific DNA-protein complex in Sf9 nuclear extracts probed with a STAT binding motif oligomer.
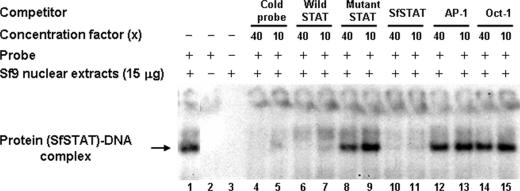
EMSA was used to confirm that endogenous Sf9 cell STAT was able to bind to the WSSV ie1 promoter STAT binding site. In this analysis, which used a radioactively labeled probe that contained the predicted STAT binding site of the WSSV ie1 promoter, EMSA detected a protein-DNA complex (i.e., endogenous Sf9 cell STAT protein complexed with the isotope-labeled DNA probe) in Sf9 cell nuclear extracts (Fig. 5). Specificity was confirmed by running the EMSA in the presence of different competitors at a 10× or 40× molar excess. The cold probe and the wild-type STAT and SfSTAT oligonucleotides all out-competed the radioactive probe, while the mutant STAT and the two transcriptional factor oligomers, AP-1 and Oct-1, all failed to displace the probe (Fig. 5).
FIG. 5.
EMSA of the −84/−64 region of the WSSV ie1 promoter with Sf9 cell nuclear extracts. Lane 1, 15 μg Sf9 cell nuclear extracts reacted with isotope-labeled probe (SfSTAT-DNA complex). Lane 2, isotope-labeled probe only. Lane 3, Sf9 cell nuclear extracts only. Lanes 4 to 15, SfSTAT-DNA complex competing with unlabeled probe, unlabeled wild type STAT oligonucleotide, 4-mer mutant STAT oligonucleotide, unlabeled SfSTAT oligonucleotide, unlabeled AP-1 oligonucleotide, and Oct-1 oligonucleotide. The relative concentrations of the unlabeled competitor oligonucleotides (40× or 10×) are indicated.
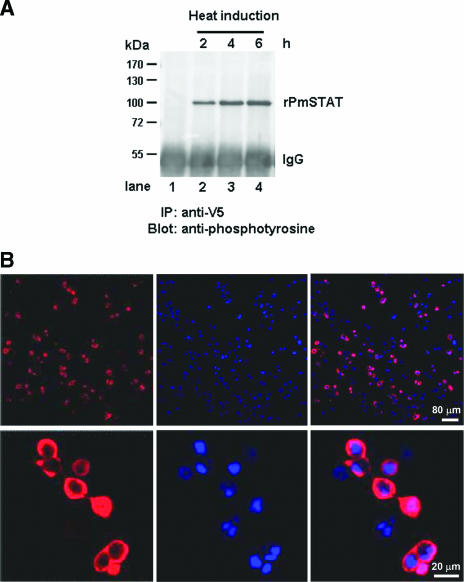
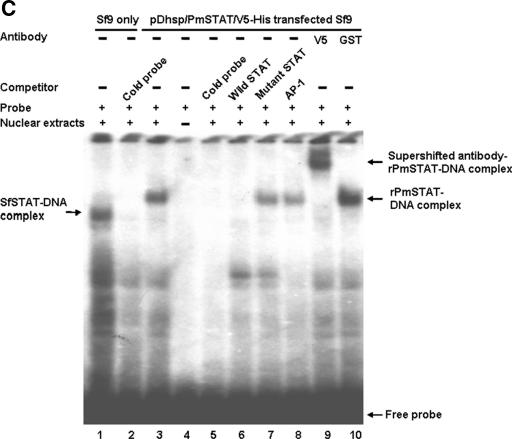
To confirm that the overexpressed full-length rPmSTAT is in an activated (i.e., phosphorylated) state even without pathogen induction, immunoprecipitation with an anti-V5 antibody followed by Western blotting with an antiphosphotyrosine antibody was performed on nuclear extracts prepared from transiently transfected Sf9 cells after heat induction. In the Sf9 cells transfected with pDhsp/PmSTAT/V5-His, the amounts of phosphorylated rPmSTAT increased with time (2 to 6 h) after heat shock induction (Fig. 6A). Cellular localization of the rPmSTAT in the pDhsp/PmSTAT/V5-His-transfected Sf9 insect cells 6 h after heat induction was predominantly in the cytoplasm, but some of the stained rPmSTAT was also seen in the nucleus (Fig. 6B). In the EMSA, nuclear extracts were prepared from pDhsp/PmSTAT/V5-His-transfected Sf9 cells at 6 h after heat induction, and an rPmSTAT-DNA probe complex was detected (Fig. 6C, lane 3). The SfSTAT-DNA band was not seen in lane 3 (or in lanes 7 to 10) because the overexpressed and activated rPmSTAT out-competed the endogenous SfSTAT. Assays in the presence of other competitors in 40× molar excess confirmed the specificity of the radiolabeled probe (Fig. 6C, lanes 5 to 8). Since the rPmSTAT plasmid was constructed by ligation to a vector that already contained the V5 epitope, probing with anti-V5 antibody resulted in a supershifted band that contained an antibody-rPmSTAT-DNA complex (Fig. 6C, lane 9). No supershifted complex was formed in a control assay with a different antibody (GST) (Fig. 6C, lane 10). These data all suggest that STAT (both SfSTAT and rPmSTAT) interacts with the putative STAT binding site located in the region from nt −84 to −64 of the WSSV ie1 promoter and that this contributes to the high WSSV ie1 promoter activity in Sf9 insect cells.
FIG.6.
(A) Immunoprecipitation (IP) and Western blotting analysis of the phosphorylation status of rPmSTAT. Lane 1 (negative control), nuclear extract from Sf9 cells transfected with the empty plasmid pDhsp/V5-His. Lanes 2 to 4, phosphorylated rPmSTAT was detected in nuclear extracts from Sf9 cells transfected with pDhsp/PmSTAT/V5-His at 2 h after heat shock induction, and quantities of (pp)rPmSTAT increased through 4 to 6 h postinduction. IgG, immunoglobulin G. (B) Immunofluorescence staining of pDhsp/PmSTAT/V5-His-transfected Sf9 cells. Cells were probed with anti-V5 antibody coupled with Cy3-labeled secondary antibody (red) to detect STAT (left column) and counterstained with DAPI (blue) to show the location of the nuclei (middle column). The merged result (right column) shows that rPmSTAT is present in the cytoplasm and sometimes in the nuclei of the Sf9 cells. (C) EMSA and supershift EMSA to confirm that the WSSV ie1 promoter putative STAT binding region is binding to STAT. Lane 1, SfSTAT-DNA complex of 32P-labeled probe and untransfected Sf9 cell nuclear extract. Lane 2, SfSTAT-DNA complex competing with unlabeled probe. Lane 3, rPmSTAT-DNA complex of labeled probe and pDhsp/PmSTAT/V5-His-transfected Sf9 cell nuclear extract. Lane 4, labeled probe only. Lane 5, rPmSTAT-DNA complex competing with unlabeled probe. Lane 6, rPmSTAT-DNA complex competing with unlabeled wild-type STAT oligonucleotide. Lane 7, rPmSTAT-DNA complex competing with unlabeled mutant STAT oligonucleotide. Lane 8, rPmSTAT-DNA complex competing with unlabeled AP-1 oligonucleotide. Lane 9, binding of rPmSTAT-DNA complex with anti-V5 antibody directed against a V5 tag in rPmSTAT. Lane 10, an anti-GST antibody not specific for rPmSTAT failed to bind with and supershift the rPmSTAT-DNA complex. The concentration of the unlabeled competitors was in 40× molar excess relative to the labeled probe.
PmSTAT is activated in response to WSSV infection in shrimp.
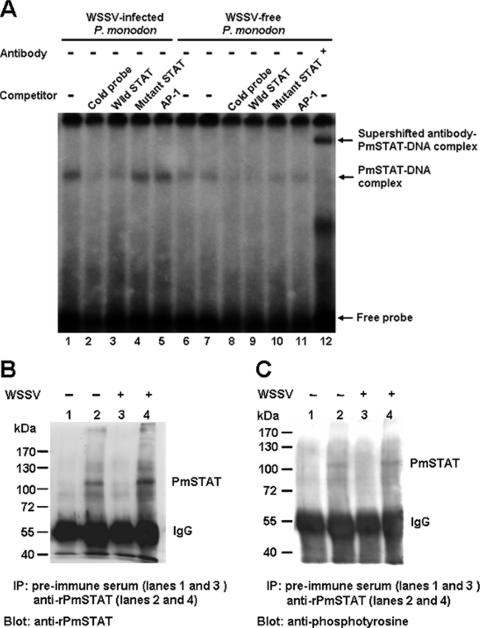
To determine whether WSSV infection would influence the activity of PmSTAT, EMSA and the same ie1 STAT binding motif oligomer probe were used to investigate the activation states of PmSTAT in nuclear extracts from WSSV-challenged and unchallenged P. monodon. As Fig. 7A shows, DNA binding activity was observed in nuclear extracts from both WSSV-infected P. monodon (72 hpi) (lane 1) and WSSV-free P. monodon (0 hpi) (lanes 6 and 7). However, the intensity of the band associated with the PmSTAT-DNA complex was higher in reactions performed on WSSV-infected P. monodon, even when the amount of WSSV-free P. monodon nuclear extract was doubled to 20 μg (lane 7). The PmSTAT-DNA complex was completely out-competed in the presence of a 40-fold excess of the unlabeled probe (lanes 2 and 8) and the wild-type STAT oligonucleotide (lanes 3 and 9), but competition with an excess of mutant STAT (lanes 4 and 10) and noncompetitor AP-1 oligomers (lanes 5 and 11) had no effect. Supershift of the DNA-protein complexes was observed in the presence of anti-rPmSTAT antibody (lane 12). Next, to compare the expression and phosphorylation levels of PmSTAT in WSSV-infected and WSSV-free P. monodon, nuclear extracts were immunoprecipitated using anti-rPmSTAT polyclonal antibody and Western blotted using either anti-rPmSTAT antibody or antiphosphotyrosine antibody. Figure 7B and C, respectively, show that in extracts from the WSSV-infected P. monodon, the expression levels of PmSTAT and the amounts of tyrosine-phosphorylated PmSTAT proteins were at least as high as those in extracts from the uninfected shrimp. Taken together, these data show that the intramuscular injection of WSSV induces PmSTAT proteins in vivo and also increases the specific DNA binding activity and tyrosine phosphorylation of these proteins.
FIG. 7.
(A) EMSAs of nuclear extracts from WSSV-infected (72 hpi) and WSSV-free P. monodon, using a 32P-labeled ie1 promoter STAT binding sequence oligonucleotide as a probe. Competitors, when present, were in a 40× molar excess relative to the “hot” probe. Lane 7 used 20 μg of nuclear extract; all the other lanes used 10 μg. The antibody used in lane 12 was specifically directed against PmSTAT. (B and C) Immunoprecipitation (IP) and Western blot analysis of PmSTAT in nuclear extracts from WSSV-infected (+) and WSSV-free (−) P. monodon. The nuclear extracts (the same as those used in the EMSA) were immunoprecipitated with anti-rPmSTAT antiserum or preimmune serum (control), separated by gel electrophoresis, and probed with either anti-rPmSTAT antibody (B) or antiphosphotyrosine antibody (C). IgG, immunoglobulin G.
Recombinant PmSTAT transactivates WSSV ie1 promoter in a dose-dependent manner.
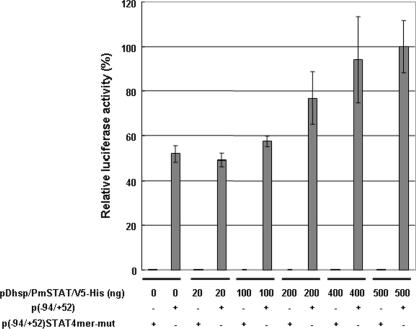
The effect of transiently expressed rPmSTAT on the luciferase activities of p(−94/+52) in Sf9 cells (Fig. 8) shows that rPmSTAT influences WSSV ie1 promoter transcriptional activity in a dose-dependent manner. When the p(−94/+52) plasmid was replaced by the ie1 STAT binding region mutant, p(−94/+52)STAT4mer-mut, the relative luciferase activity fell to almost zero.
FIG. 8.
Dose-dependent transactivation of the WSSV ie1 promoter p(−94/+52) by recombinant PmSTAT. The data show relative luciferase activities at 6 h after heat shock in Sf9 cells that were cotransfected for 48 h with different concentrations of pDhsp/PmSTAT/V5-His and with either 500 ng of p(−94/+52) reporter plasmid or 500 ng of the 4-mer mutant plasmid. The relative luciferase activity of 500 ng of pDhsp/PmSTAT/V5-His was arbitrarily set to 100%. The means and standard deviations from three independent transfections are shown.
DISCUSSION
In our previous study, the WSSV ie1 promoter demonstrated strong activity (24). In the present study, the WSSV ie1 promoter region was cloned and the cis elements in the region involved in the regulation of this gene were identified. The progressive deletion and mutant analyses (Fig. 1, 2, 3, and 4) showed that the basal promoter was located between nt −71 and +52, which is a region that includes the TATA box and transcriptional start site (24), and also suggested that a 23-mer fragment from nt −94 to −72 was critical for the promoter's strong activity. This element was not an enhancer, but it formed part of a STAT binding sequence (Fig. 4A). This STAT binding motif acted as a transcriptional factor binding site that was transactivated by endogenous SfSTAT (Fig. 5), by recombinant PmSTAT (Fig. 6), and by endogenous PmSTAT (Fig. 7), and it accounted for the strong ie1 promoter activity. Experiments performed with Sf9 insect cells further showed that WSSV ie1 promoter was transactivated by rPmSTAT in a dose-dependent manner (Fig. 8). However, in Fig. 8, the ie1 promoter shows relatively high activities even in the cells that were cotransfected with the mock plasmid pDhsp/V5-His, and as the amount of rPmSTAT was increased to 500 ng, ie1 promoter activity increased by only a factor of 2. The high activity levels even in the absence of PmSTAT are presumably due to the presence of the Sf9 cells' endogenous STATs. Meanwhile, the site-mutated reporter plasmid p(−94/+52)STAT4mer-mut was not activated either by the endogenous SfSTAT or by the recombinant PmSTAT, because its mutated STAT binding site presumably prevents it from binding.
PmSTAT is a 774-amino-acid protein that contains a protein interaction domain, a DNA binding domain, an SH2 domain, and a single C-terminal tyrosine residue, all of which are commonly found in all STAT-like genes (19). In Sf9 cells, two STATs have been identified (GenBank accession no. AF329946 and AF329947), but they differ only in the 3′ ends of their coding regions. The putative amino acid sequences of PmSTAT show ∼65% similarity and ∼50% identity, respectively, to these two SfSTATs, and they also share 88% similarity and 80% identity in the DNA binding domain. These data are consistent with the idea that the STAT binding motif of the WSSV ie1 promoter is recognized and activated by both the P. monodon and the Sf9 STATs. The EMSA findings in Fig. 5 confirmed the binding of endogenous SfSTAT to the WSSV ie1 promoter. Lane 1 of Fig. 6C also shows that endogenous SfSTAT binds to the ie1 promoter in Sf9 cells and that it is out-competed by the overexpressed active form of rPmSTAT (cf. lanes 3, 7, 8, 9, and 10, where the SfSTAT-DNA band is absent). All these data suggest that SfSTAT and PmSTAT may be somewhat similar in function, even if these functions remain unclear. At the very least, though, we have shown here that both SfSTAT and PmSTAT are capable of transactivating WSSV ie1 gene expression.
STATs represent a family of latent transcription factors that are activated upon tyrosine phosphorylation in response to extracellular signals (3, 9, 45). Tyrosine-phosphorylated STATs form dimers or multimers, are transported into the nucleus, bind to the cognate DNA sequences, and activate gene expression (7, 10, 15, 31, 44). In vertebrates, the antiviral function of STATs is well known (30, 36), but more recently, the JAK-STAT signaling pathway has also been shown to be involved in the antiviral response of Drosophila infected with Drosophila C virus (11). STATs may also be part of the antiviral response in a mosquito (Aedes albopictus) cell line, but in this case, when the cell line is infected with Japanese encephalitis virus, the virus acts to reduce the DNA binding activity and tyrosine phosphorylation of the cellular STAT (23). In human, instead of inhibiting STAT activity, some viruses annex host STATs to the apparent advantage of the virus. For example, during infection with Kaposi's sarcoma-associated herpesvirus, the immediate-early gene product ORF50 induces STAT3 translocation into the nucleus, where it binds to and stabilizes STAT3 dimer to increase the oncogenic potential of the infected cells (13). Similarity, the enhancer element in hepatitis B virus (HBV) regulates the organ-specific activity of the HBV promoter elements (4, 14, 18, 32), and in HBV-infected hepatocytes, activation of STAT3 combined with another factor (hepatocyte nuclear factor 3) leads to the activation of the enhancer element 1 function that controls HBV gene expression (41). The present study now shows that a similar phenomenon also occurs in a crustacean; that is, during infection with WSSV, a crustacean STAT is annexed to the apparent benefit of the virus. Furthermore, this binding of PmSTAT to the ie1 promoter is seen not just in the Sf9 insect cell line but also in vivo in P. monodon (Fig. 7). The replication strategy of WSSV in its crustacean hosts might therefore resemble HBV's strategy in liver. We conclude that, like HBV, WSSV does not inhibit the presumably defensive STAT activity of its host, but on the contrary, WSSV annexes the host STAT and uses it to activate viral gene expression.
Acknowledgments
This investigation was supported financially by National Science Council grants (NSC 94-2317-B-002 -010 and NSC 94-2317-B-002 -011).
We are indebted to Paul Barlow for his helpful criticism.
Footnotes
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Agaisse, H., and N. Perrimon. 2004. The roles of JAK/STAT signaling in Drosophila immune responses. Immunol. Rev. 198:72-82. [DOI] [PubMed] [Google Scholar]
- 2.Agaisse, H., U. M. Petersen, M. Boutros, B. Mathey-Prevot, and N. Perrimon. 2003. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev. Cell. 5:441-450. [DOI] [PubMed] [Google Scholar]
- 3.Akira, S., Y. Nishio, M. Inoue, X. J. Wang, S. Wei, T. Matsusaka, K. Yoshida, T. Sudo, M. Naruto, and T. Kishimoto. 1994. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell 77:63-71. [DOI] [PubMed] [Google Scholar]
- 4.Antonucci, T. K., and W. J. Rutter. 1989. Hepatitis B virus (HBV) promoters are regulated by the HBV enhancer in a tissue-specific manner. J. Virol. 63:579-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barillas-Mury, C., Y.-S. Han, D. Seeley, and F. C. Kafatos. 1999. Anopheles gambiae Ag-STAT, a new insect member of the STAT family, is activated in response to bacterial infection. EMBO J. 18:959-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blissard, G. W. 1996. Baculovirus-insect cell interactions. Cytotechnology 20:73-93. [DOI] [PubMed] [Google Scholar]
- 7.Chen, X., U. Vinkemeier, Y. Zhao, D. Jeruzalmi, J. E. Darnell, Jr., and J. Kuriyan. 1998. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell 93:827-839. [DOI] [PubMed] [Google Scholar]
- 8.Chou, H.-Y., C.-Y. Huang, C.-H. Wang, H.-C. Chiang, and C.-F. Lo. 1995. Pathogenicity of a baculovirus infection causing white spot syndrome in cultured penaeid shrimp in Taiwan. Dis. Aquat. Org. 23:165-173. [Google Scholar]
- 9.Darnell, J. E., Jr. 1997. STATs and gene regulation. Science 277:1630-1635. [DOI] [PubMed] [Google Scholar]
- 10.Darnell, J. E., Jr., I. M. Kerr, and G. M. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 11.Dostert, C., E. Jouanguy, P. Irving, L. Troxler, D. Galiana-Arnoux, C. Hetru, J. A. Hoffmann, and J. L. Imler. 2005. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat. Immunol. 6:946-953. [DOI] [PubMed] [Google Scholar]
- 12.Flegel, T. W. 1997. Major viral disease of the black tiger prawn (Penaeus monodon) in Thailand. World J. Microbiol. Biotechnol. 13:433-442. [Google Scholar]
- 13.Gwack, Y., S. Hwang, C. Lim, Y. S. Won, C. H. Lee, and J. Choe. 2002. Kaposi's sarcoma-associated herpesvirus open reading frame 50 stimulates the transcriptional activity of STAT3. J. Biol. Chem. 277:6438-6442. [DOI] [PubMed] [Google Scholar]
- 14.Honigwachs, J., O. Faktor, R. Dikstein, Y. Shaul, and O. Laub. 1989. Liver-specific expression of hepatitis B virus is determined by the combined action of the core gene promoter and the enhancer. J. Virol. 63:919-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horvath, C. M., Z. Wen, and J. E. Darnell, Jr. 1995. A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 9:984-994. [DOI] [PubMed] [Google Scholar]
- 16.Hossain, M. S., S. Khadijah, and J. Kwang. 2004. Characterization of ORF89—a latency-related gene of white spot syndrome virus. Virology 325:106-115. [DOI] [PubMed] [Google Scholar]
- 17.Inouye, K., S. Miwa, N. Oseko, H. Nakano, and T. Kimura. 1994. Mass mortalities of cultured kuruma shrimp, Penaeus japonicus, in Japan in 1993: electron microscope evidence of the causative virus. Fish Pathol. 29:149-158. (In Japanese.) [Google Scholar]
- 18.Jameel, S., and A. Siddiqui. 1986. The human hepatitis B virus enhancer requires trans-acting cellular factor(s) for activity. Mol. Cell. Biol. 6:710-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kisseleva, T., S. Bhattacharya, J. Braunstein, and C. W. Schindler. 2002. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene 285:1-24. [DOI] [PubMed] [Google Scholar]
- 20.Kwon, E. J., H. S. Park, Y. S. Kim, E. J. Oh, Y. Nishida, A. Matsukage, M. A. Yoo, and M. Yamaguchi. 2000. Transcriptional regulation of the Drosophila raf proto-oncogene by Drosophila STAT during development and in immune response. J. Biol. Chem. 275:19824-19830. [DOI] [PubMed] [Google Scholar]
- 21.Lahiri, D. K., and Y.-W. Ge. 2000. Electrophoretic mobility shift assay for the detection of specific DNA-protein complex in nuclear extracts from the cultured cells and frozen autopsy human brain tissue. Brain Res. Protoc. 5:257-265. [DOI] [PubMed] [Google Scholar]
- 22.Leu, J.-H., J.-M. Tsai, H.-C. Wang, A.-H. Wang, C.-H. Wang, G.-H. Kou, and C.-F. Lo. 2005. The unique stacked rings in the nucleocapsid of the white spot syndrome virus virion are formed by the major structural protein VP664, the largest viral structural protein ever found. J. Virol. 79:140-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, C.-C., C.-M. Chou, Y.-L. Hsu, J.-C. Lien, Y.-M. Wang, S.-T. Chen, S.-C. Tsai, P.-W. Hsiao, and C.-J. Huang. 2004. Characterization of two mosquito STATs, AaSTAT and CtSTAT. J. Biol. Chem. 279:3308-3317. [DOI] [PubMed] [Google Scholar]
- 24.Liu, W.-J., Y.-S. Chang, C.-H. Wang, G.-H. Kou, and C.-F. Lo. 2005. Microarray and RT-PCR screening for white spot syndrome virus immediate-early genes in cycloheximide-treated shrimp. Virology 334:327-341. [DOI] [PubMed] [Google Scholar]
- 25.Lo, C.-F., C.-H. Ho, C.-H. Chen, K.-F. Liu, Y.-L. Chiu, P.-Y. Yeh, S.-E. Peng, H.-C. Hsu, H.-C. Liu, C.-F. Chang, M.-S. Su, C.-H. Wang, and G.-H. Kou. 1997. Detection and tissue tropism of white spot syndrome baculovirus (WSBV) in captured brooders of Penaeus monodon with a special emphasis on reproductive organs. Dis. Aquat. Org. 30:53-72. [Google Scholar]
- 26.Lo, C.-F., C.-H. Ho, S.-E. Peng, C.-H. Chen, H.-C. Hsu, Y.-L. Chiu, C.-F. Chang, K.-F. Liu, M.-S. Su, C.-H. Wang, and G.-H. Kou. 1996. White spot syndrome baculovirus (WSBV) detected in cultured and captured shrimp, crabs and other arthropods. Dis. Aquat. Org. 27:215-225. [Google Scholar]
- 27.Lu, L., H. Wang, I. Manopo, L. Yu, and J. Kwang. 23. August 2005, posting date. Baculovirus-mediated promoter assay and transcriptional analysis of white spot syndrome virus orf427 gene. Virol. J. 2. http://www.virologyj.com/content/2/1/71. [DOI] [PMC free article] [PubMed]
- 28.Momoyama, K., M. Hiraoka, H. Nakano, H. Koube, K. Inouye, and N. Oseka. 1994. Mass mortalities of cultured kuruma shrimp, Penaeus japonicus, in Japan in 1993: histopathological study. Fish Pathol. 29:141-148. [Google Scholar]
- 29.Nakano, H., H. Koube, S. Umezawa, K. Momoyama, M. Hiraoka, K. Inouye, and N. Oseko. 1994. Mass mortalities of cultured kuruma shrimp, Penaeus japonicus, in Japan in 1993: epizootiological survey and infection trails. Fish Pathol. 29:135-139. [Google Scholar]
- 30.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schindler, C., and J. E. Darnell, Jr. 1995. Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu. Rev. Biochem. 64:621-651. [DOI] [PubMed] [Google Scholar]
- 32.Shaul, Y., W. J. Rutter, and O. Laub. 1985. A human hepatitis B enhancer element. EMBO J. 4:427-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sliva, D., and L.-A. Haldosen. 1996. STAT-like DNA-binding activity in Spodoptera frugiperda cells. Biochem. Biophys. Res. Commun. 225:562-569. [DOI] [PubMed] [Google Scholar]
- 34.Sliva, D., T. J. Wood, C. Schindler, P. E. Lobie, and G. Norstedt. 1994. Growth hormone specifically regulates serine protease inhibitor gene transcription via gamma-activated sequence-like DNA elements. J. Biol. Chem. 269:26208-26214. [PubMed] [Google Scholar]
- 35.Tacchini, L., D. Fusar-Poli, and A. Bernelli-Zazzera. 2002. Activation of transcription factors by drugs inducing oxidative stress in rat liver. Biochem. Pharmacol. 63:139-148. [DOI] [PubMed] [Google Scholar]
- 36.Takaoka, A., and H. Yanai. 2006. Interferon signalling network in innate defence. Cell. Microbiol. 8:907-922. [DOI] [PubMed] [Google Scholar]
- 37.Torok, I., and F. Karch. 1980. Nucleotide sequences of heat shock activated genes in Drosophila melanogaster. I. Sequences in the regions of the 5′ and 3′ ends of the hsp 70 gene in the hybrid plasmid 56H8. Nucleic Acids Res. 8:3105-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai, J.-M., H.-C. Wang, J.-H. Leu, A. H.-J. Wang, Y. Zhuang, P. J. Walker, G.-H. Kou, and C.-F. Lo. 2006. Identification of the nucleocapsid, tegument, and envelope proteins of the shrimp white spot syndrome virus virion. J. Virol. 80:3021-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai, J.-M., H.-C. Wang, J.-H. Leu, H.-H. Hsiao, A. H.-J. Wang, G.-H. Kou, and C.-F. Lo. 2004. Genomic and proteomic analysis of thirty-nine structural proteins of shrimp white spot syndrome virus. J. Virol. 78:11360-11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vlak, J. M., J. R. Bonami, T. W. Flegel, G. H. Kou, D. V. Lightner, C.-F. Lo, P. C. Loh, and P. W. Walker. 2004. Nimaviridae, p. 187-192. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: eight report of the International Committee on Taxonomy of Viruses. Elsevier, Philadelphia, PA.
- 41.Waris, G., and A. Siddiqui. 2002. Interaction between STAT-3 and HNF-3 leads to the activation of liver-specific hepatitis B virus enhancer 1 function. J. Virol. 76:2721-27299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wongteerasupaya, C., J. E. Vickers, S. Sriurairatana, G. L. Nash, A. Akarajamorn, V. Boonsaeng, S. Panyim, A. Tassanakajon, B. Withyachumnarnkul, and T. W. Flegel. 1995. A non-occluded, systemic baculovirus that occurs in cells of ectodermal and mesodermal origin and causes high mortality in the black tiger prawn Penaeus monodon. Dis. Aquat. Org. 21:69-77. [Google Scholar]
- 43.Xie, X., L. Xu, and F. Yang. 2006. Proteomic analysis of the major envelope and nucleocapsid proteins of white spot syndrome virus. J. Virol. 80:10615-10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu, X., Y. L. Sun, and T. Hoey. 1996. Cooperative DNA binding and sequence-selection recognition conferred by the STAT amino-terminal domain. Science 273:794-797. [DOI] [PubMed] [Google Scholar]
- 45.Zhong, Z., Z. Wen, and J. E. Darnell, Jr. 1994. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 264:95-98. [DOI] [PubMed] [Google Scholar]