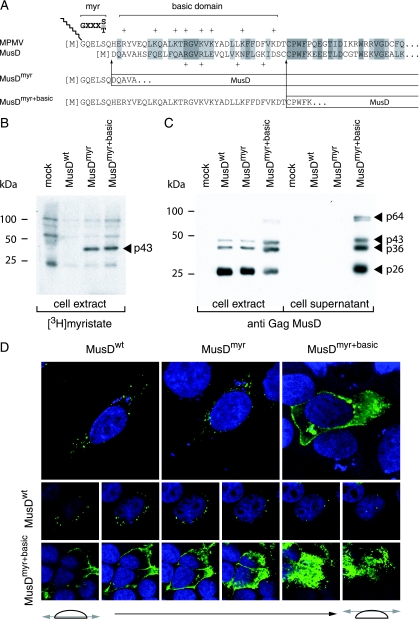
FIG. 2.
Consequences of restoring myristoylation and membrane targeting signals on MusD Gag protein addressing. (A) Sequence of the N-terminal domain of the MusD Gag protein and comparison with the corresponding domain from its closest relative, the type-D MPMV retrovirus. The consensus sequence required for myristoylation (myr, [M]GXXXS/T) and the MPMV domain rich in basic residues (basic domain, +) are indicated. Arrows indicate the limits of the replacements between the N-terminal residues of MPMV and MusD Gag proteins to generate the MusDmyr and MusDmyr+basic proviruses, respectively. (B) Restoration of MusD Gag myristoylation. Whole-cell lysates were prepared from human 293T cells transfected with the indicated MusD constructs and labeled with [3H]myristate. Myristoylated proteins were revealed by fluorography after separation by SDS-PAGE. (C) Western blot analysis of whole-cell lysates or cell supernatants from 293T cells transfected as in panel B with the indicated MusD constructs, using a rabbit antiserum directed against the MusD Gag polyprotein. The molecular masses of the Gag-specific bands are indicated in kilodaltons, as in Fig. 1E. (D) Immunofluorescence confocal analysis of human HeLa cells transfected with the indicated MusD constructs. Cells were grown on glass coverslips approximately 48 h posttransfection, fixed, and permeabilized, and MusD Gag proteins were detected by using the same rabbit anti-Gag MusD antiserum as in panel C and an Alexa Fluor 488-conjugated anti-rabbit secondary antibody. Nuclei were stained with TO-PRO-3 iodide. The two rows at the bottom show the successive confocal images of stained cells.