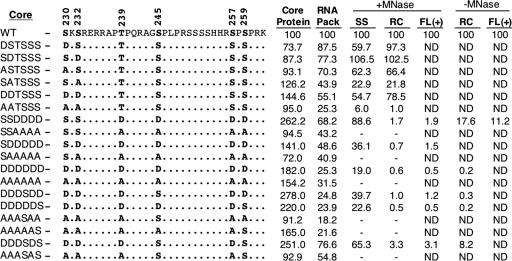
FIG. 1.
Summary of DHBV core phosphorylation mutants. The sequences of the C-terminal 33 amino acids of the WT and various mutants are shown. The S or T residues at the six known phosphorylation sites (shown in bold) were changed to either A or D, as indicated. The mutants are designated on the left. The average levels in the various core mutants, compared to those in the WT (set to 100), of core protein expression (as measured by SDS-PAGE and Western blot analysis), pgRNA packaging (RNA pack) (as measured by the native agarose gel electrophoresis assay), SS DNA (extracted with micrococcal nuclease [MNase] digestion), RC DNA (extracted with or without exogenous nuclease digestion), and full-length plus-strand DNA [FL (+), extracted with or without nuclease digestion and measured using denatured core DNA], are indicated. ND, not determined; −, levels of DNA were too low to be quantified reliably.