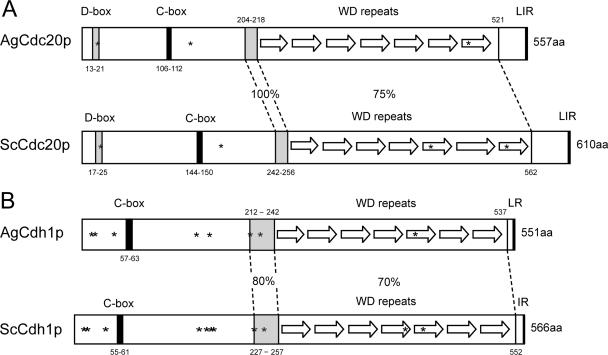
FIG. 1.
Comparison of homologous domains and sequence features in APC/C cofactors from A. gossypii and S. cerevisiae. (A) AgCdc20p (AFL014C) and ScCdc20p (YGL116W) are 59% identical on the amino acid level. The positions of a D box, a C box, and seven WD repeats are shown for each protein, and notably, a stretch of 15 identical amino acids (aa) precedes the WD repeats. Asterisks mark potential CDK phosphorylation sites (S20, S136, and S493 in AgCdc20p and S24, S173, S439, and S534 in ScCdc20p). These sites are conserved in eight yeast species and A. gossypii except S439 (in the fifth WD repeat). In all nine Cdc20p orthologues, the last amino acid is arginine preceded by two hydrophobic amino acids. (B) AgCdh1p (AFL007C) and ScCdh1p (YGL003C) are 66% identical, with the highest homology in the WD repeat region and the preceding 21 amino acids. All nine analyzed Cdh1 orthologues terminate with an arginine preceded by a hydrophobic amino acid. The eight asterisks in AgCdh1p mark CDK phosphoryation consensus sites (T12, S16, S44, T144, T161, S212, S224, and S421), all of which are conserved in eight yeast orthologues. In ScCdh1p, the homologous sites are phosphorylated with the possible exceptions of S42 and S227 (16a, 57). The three additional consensus CDK sites in ScCdh1p (S169, T173, and S418) are most likely not phosphorylated. From the 11 non-CDK phosphorylation sites identified in ScCdh1p by those authors, only 5 (S38, S172, S193, S225, and S556) are conserved in A. gossypii and other yeast Cdh1 sequences. LIR, LR, and IR refer to amino acids.