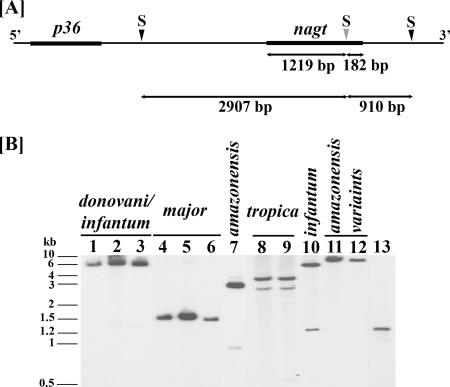
FIG. 3.
Leishmania nagt gene as a single-copy gene in representative genotypes. (A) Genomic SacII (S) map of the L. amazonensis nagt gene and its flanking regions. Note a single intragenic SacII site (true for all nagt genotypes of the Leishmania subgenus) near the 3′ end of the nagt ORF (thick line) at nt position 1219 (gray arrow head) and two flanking sites (black arrow head) [placed on the basis of sequences known for this chromosomal region of L. amazonensis (36)] at ∼2.9 kb upstream and ∼0.9 kb downstream of the intragenic SacII site. The p36 gene is another ORF ∼2.3 kb upstream of nagt, shown here for orientation. (B) Southern blot analysis of SacII-restricted genomic DNAs from representative Leishmania genotypes showing nagt as a single-copy gene. SacII digests of genomic DNAs were run in 0.8% agarose gel for Southern blotting analyses with the PCR-amplified L. amazonensis nagt gene as the probe. All samples gave at least one strong signal after a short exposure and no more than two after prolonged exposure (not shown). Note that the small variation among different samples in signal intensity is due to slightly unequal loading of the genomic DNAs. The two positive SacII fragments of the L. amazonensis nagt gene are exactly as expected in size and in intensity as mapped (panel A) for genomic DNA (lane 7) and for PCR products (lane 13). Lanes 1 to 3, L. donovani, L. infantum variant 7, and L. chagasi (infantum); lanes 4 to 6, L. major variant 1, L. major variant 4, and L. major; lane 7, L. amazonensis; lanes 8 to 9, L. tropica (I/II); lane 10, L. infantum; lanes 11 and 12, L. amazonensis variant 17 and L. amazonensis variant 6; lane 13, PCR-amplified L. amazonensis nagt gene. See Table 1 for L. amazonensis, L. infantum, and L. donovani designated as genotypes 1, 5, and 9, respectively.