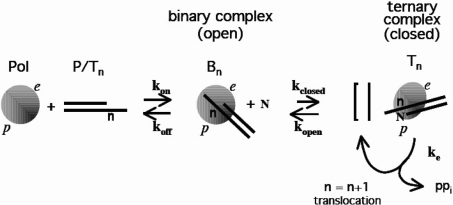
Figure 7. Model for DNA replication by T4 polymerase based on structural data for the homologous DNA polymerase RB69 [28–32].
In this scheme T4 DNA polymerase (Pol), which has a polymerase catalytic site (p) and an exonuclease catalytic site (e), binds to an undamaged P/T DNA with base n at the coding position on the template strand (P/Tn) to form a binary complex (Bn). Pol and Bn are in an open conformation indicated as a sphere and P/Tn is situated in the exonuclease site of the binary complex (see text). Binding of complementary substrate dNTP (N) to Bn induces a closed ternary complex (Tn) depicted as an ellipse. During this conformational change the P/T DNA rotates 40 ° and its extremity moves 40 Å (1 Å=0.1 nm) so that N, n and the 3′-OH terminus of the primer strand are at p and the enzyme catalyses polymerization (ke). Addition of N to the primer strand is followed by pyrophosphate (PPi) release and translocation. Subsequent nucleotide addition that leads to elongation probably occurs via a closed polymerase complex, designated [], whose structure has not yet been determined. The polymerase complex attains thermodynamic equilibrium between each nucleotide addition step [36] and consequently can either dissociate or bind the following dNTP and enter the next elongation cycle.