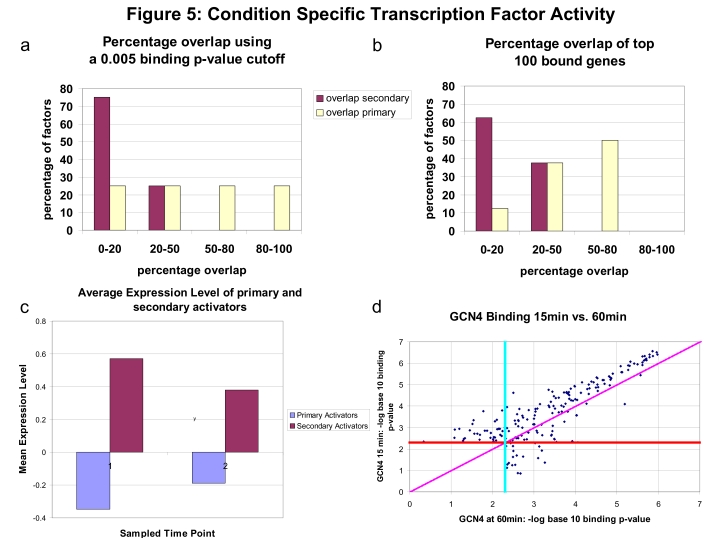
Figure 5.
Distribution of binding overlap between YPD media and stress condition for different sets of TFs. We group TF into two sets. The first subset contains the TFs assigned to first split points in AA starvation or heat that were profiled with a ChIP-chip experiment in the condition (8, primary TFs Cbf1, Gcn4, Fhl1, Rap1, and Sfp1 from AA starvation in Harbison et al (2004) and for heat Msn2 and Skn7 from Harbison et al (2004) and Hsf1 from Hahn et al (2004)) and the secondary TFs assigned to second split point profiled with a ChIP-chip experiment in the condition (8, secondary TFs Arg81, Dal82, Gln3, Hap5, Met32, Met4, Rtg3, and Stp1 from AA starvation in Harbison et al, 2004). (A) Percent overlap for each of these two sets when binding (under both stress and YPD media conditions) is determined using a 0.005 P-value cutoff. Note the difference between the distribution of overlap for primary and other TFs. Whereas the majority of TFs display a big difference in the set bound genes under stress and YPD media, many primary TFs bind to a large percentage of stress-regulated genes in YPD media as well. This difference is even bigger in (B) where we plot the overlap for the top 100 genes (ordered by their binding P-values) in each condition. Whereas most TFs (and most secondary TFs) drastically alter the subset of genes they regulate under stress, half of the primary factors bind to more than 50% of the same genes in both conditions. Although the binding strength may be different under stress, these results indicate that many of the primary pathways are maintained, in low levels, under YPD media as well. (C) Average expression levels for primary and secondary factors for the first two time points in the AA starvation and heat-shock experiments. Whereas the average expression levels for the secondary factors are much higher when compared to their untreated levels, the levels for the primary factors do not change significantly between the two conditions. (D) Comparison of binding P-values for Gcn4 in the MMS condition at 15 and 60 min. Points above the diagonal correspond to genes that were bound more significantly at 15 min than at 60 min. As can be seen from the plot, a substantial majority of genes were bound more significantly at 15 min than at 60 min. Points to the right of the vertical had a P-value <0.005 in MMS at 60 min, whereas points above the horizontal line had a P-value <0.005 in MMS at 15 min.