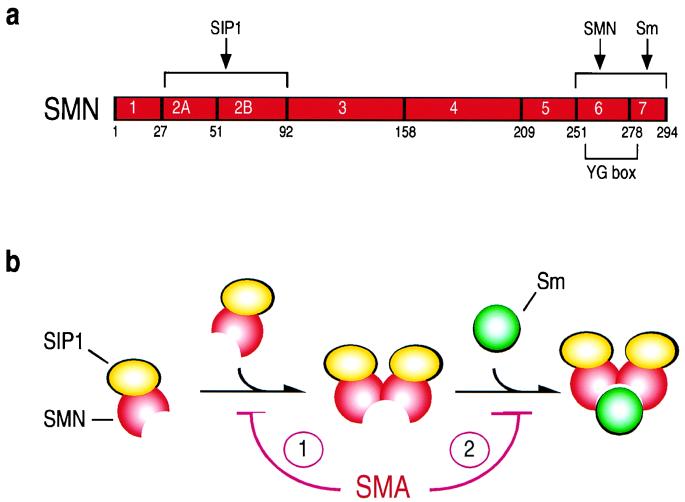
Figure 6.
Schematic model depicting the interaction of SMN/SIP1 with Sm proteins. (a) Schematic structure of the SMN protein and its interacting domain. The amino acid numbers and the borders of exons are indicated. SIP1-interacting domain resides at the amino terminus of SMN as determined by competition experiments (10). SMN self-association and SMN/Sm interaction domains overlap with the conserved YG box at the carboxyl-terminus of SMN as determined by deletion, mutation, and competition experiments (refs. 10 and 11 and this paper). (b) Monomeric SMN, associated with SIP1, which binds to SMN but not to itself (ref. 10; data not shown), contains a low-affinity binding site for Sm proteins. SMN self-associates, forming at least a SMN/SIP1 tetrameric complex. In this oligomeric conformation a binding site is formed with a much higher affinity for the Sm proteins. SMN mutations found in SMA patients result in a reduced ability of SMN to self-associate (1) and also map within the Sm-binding site itself (2), thus affecting the SMN interaction with Sm proteins.