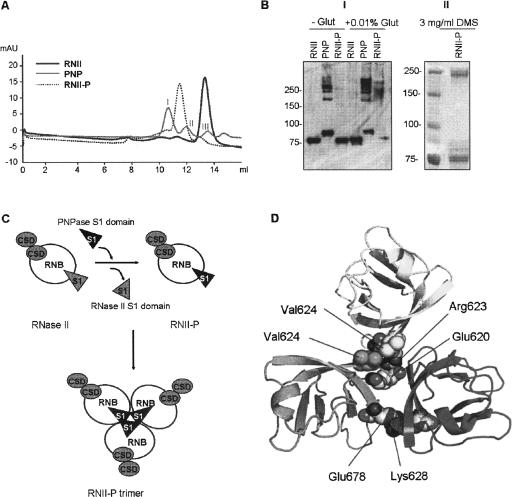
FIGURE 4.
(A) Gel filtration of (His)6–RNase II (RNII), (His)6–PNPase (PNP), and the hybrid protein (His)6–RNase II-P (RNII-P). (B) Panel I: Western blot of cross-linking experiments of (His)6–RNase II (RNII), (His)6–PNPase (PNP), and (His)6–RNase II-P (RNII-P). Samples of 0.5 μg of each protein were incubated in the absence (−) or in the presence (+) of 0.01% of gluteraldehyde and analyzed in a SDS-10% PAGE gel. Standard molecular weight is indicated on the left; Panel II: Western blot of cross-linking experiments of (His)6–RNase II-P (RNII-P). Ten micrograms of the protein were incubated in the presence of 3 mg/mL of dimethy suberimidate and analyzed in a SDS-10% PAGE gel. Standard molecular weight is indicated on the left. (C) Schematic representation of the model of RNase II trimerization induced by the S1 domain from PNPase. Catalytic RNB domain is represented as white circles, CSD domains are shown as gray circles, and S1 domains as triangles. (D) Detail of modeled PNPase S1 domain homotrimerization. Position of residues implicated in the two main S1 domain–S1 domain interactions in terms of stabilization of homotrimer are represented as spheres. Acidic chains of Glu 620 and Glu 678 of each monomer are faced to the side chains of positive residues Lys 628 and Arg 623, respectively, of the other two monomers. Contacts among side chains of all three Val 624 residues configure a local hydrophobic cluster. Only one representative of both interactions is depicted for clarity.