TABLE 2.
Summary of simulation assumptions
| Single-locus model RL | Multilocus model P |
|---|---|
| Genotypic structure | |
| One disease-susceptibility locus on one chromosome. There are one wild-type (N) allele and k − 1 disease (S) alleles. | Multiple (L) disease susceptibility loci on different chromosomes, with the same maximum number of allelic states (k). |
| Selection model | |
| Fitness of an individual follows either an additive or a recessive model. Fitness of an individual with genotype NN, NS, or SS is 1, 1 − s/2, 1 − s for an additive model and 1, 1, 1 − s for a recessive model. | Each DSL follows either an additive or a recessive fitness model as in the case of the single-locus model. The overall fitness of an individual with fitness gi, i = 1,…, L at each DSL follows either a multiplicative (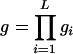 ) or an additive ( ) or an additive ( ) multilocus model. ) multilocus model. |
| Mutation model | |
k-allele (Jukes and Cantor 1969) model with k > 105 to approximate the infinite allele model. Given mutation rate μ, an allele will mutate to any other state with equal probability 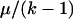 , regardless of its current allelic state. , regardless of its current allelic state. |
k-allele model with smaller k (e.g., k = 200) to be closer to a bidirectional mutation model (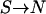 and and 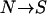 ). The mutation rate may vary from locus to locus. ). The mutation rate may vary from locus to locus. |
| Recombination | |
| NA | NA because all DSL are physically unlinked. |
| Demographic | |
| Instant, linear, or exponential population growth model. N0 = 104, N1 = 106 or 107. | Constant population size at N = 104 or 105. |
| Population structure | |
| Population may be split into equally sized subpopulations (m = 10 or 100) before population expansion. During population expansion, these subpopulations may evolve with or without migration. | NA |
| Migration | |
| Cyclic stepping-stone model at rate 10−3. | NA |
| Penetrance | |
| Indirectly modeled, see explanation in the text. | Indirectly modeled, see explanation in the text. |
