Abstract
Mutations of the methylated DNA binding protein MeCP2, a multifunctional protein that is thought to transmit epigenetic information encoded as methylated CpG dinucleotides to the transcriptional machinery, give rise to the debilitating neurodevelopmental disease Rett syndrome (RTT). In this in vitro study, the methylation-dependent and -independent interactions of wild-type and mutant human MeCP2 with defined DNA and chromatin substrates were investigated. A combination of electrophoretic mobility shift assays and visualization by electron microscopy made it possible to understand the different conformational changes underlying the gel shifts. MeCP2 is shown to have, in addition to its well-established methylated DNA binding domain, a methylation-independent DNA binding site (or sites) in the first 294 residues, while the C-terminal portion of MeCP2 (residues 295 to 486) contains one or more essential chromatin interaction regions. All of the RTT-inducing mutants tested were quantitatively bound to chromatin under our conditions, but those that tend to be associated with the more severe RTT symptoms failed to induce the extensive compaction observed with wild-type MeCP2. Two modes of MeCP2-driven compaction were observed, one promoting nucleosome clustering and the other forming DNA-MeCP2-DNA complexes. MeCP2 binding to DNA and chromatin involves a number of different molecular interactions, some of which result in compaction and oligomerization. The multifunctional roles of MeCP2 may be reflected in these different interactions.
It is now well established that the severe neurodevelopmental Rett syndrome (RTT) is caused primarily by mutations in the X-linked MeCP2 gene (1). MeCP2 (Fig. 1A) is a member of the family of related proteins that bind specifically to symmetrically methylated CpG dinucleotides via a conserved methyl binding domain (MBD) (17, 38, 44). The binding of MeCP2 to methylated DNA has been shown to lead to transcriptional repression in a variety of experimental contexts (see, for example, references 13, 35, 44, 45, and 65), a property conferred by a transcriptional repression domain (TRD). Evidence suggests that repression occurs when Sin3A and histone deacetylases (HDACs) are recruited to the TRD, resulting in the deacetylation of nearby nucleosomes (reviewed in reference 48). Additional “AT hook” and “WW” motifs have been identified in MeCP2 (9, 34), as well as a nuclear localization signal (NLS). MeCP2 is widespread and highly conserved in vertebrates, and mice lacking MeCP2 or with a major C-terminal truncation exhibit neurological dysfunctions with remarkable parallels in human RTT patients (12, 22, 49).
FIG. 1.
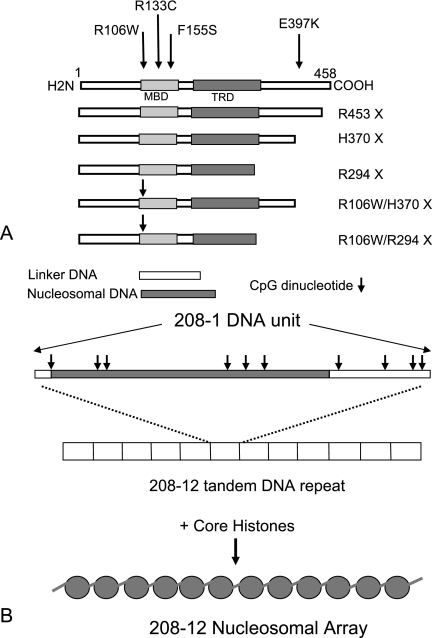
(A) Diagram of human MeCP2 with its defined functional regions, and locations of mutations and constructs used in the present study. (B) Diagram of the [208]1 DNA sequence with scheme for creating [208]12 nucleosomal arrays. The location of the preferential site for histone cores occupation is shaded, and the positions of CpG units are marked with arrows.
Analysis of RTT patients has revealed a small number of single amino acid changes at mutational “hot spots” in MeCP2, many of which are located in the MBD or TRD, as well as a series of C-terminal truncations. In addition to the hot spots, there are a large number of low-frequency mutations that lead to RTT (see the IRSA database at http://mecp2.chw.edu.au/mecp2/). There is growing evidence that MeCP2 has several functions (reviewed in references 7 and 36) but, despite considerable work, the details of the molecular interactions involved and the specific defects leading to RTT have been difficult to delineate. Transcriptional profiling indicates that MeCP2 is not a general transcriptional repressor in vivo but has a more subtle effect involving a subset of genes (3, 57). Also, the finding that MeCP2-induced repression is only partially alleviated by inhibiting HDACs (64) suggests that its activity is not restricted to HDAC recruitment. Further, there is evidence that MeCP2 is responsible for the formation of large chromatin loops (26) and involved in the regulation of RNA splicing (63). MeCP2 bound to methylated DNA may also play a regulatory role unrelated to RTT in cancer cells from breast (43, 50, 56), lung (55), and leukemias (4). MeCP2 also appears to be important in the growth of prostate cancer (6).
Extensive studies of the binding of MeCP2 (or the MBD alone) to DNA in vitro have revealed that the affinity for methylated DNA is not strong (2) and is only ∼3-fold weaker for unmethylated DNA (15). Recognizing that the in vivo substrate for MeCP2 is chromatin rather than naked DNA, we recently examined the interaction between MeCP2 and defined nucleosomal arrays (NAs) (20). This study revealed that MeCP2 induced a striking compaction of the arrays, suggesting that a change in conformation of this sort may contribute to MeCP2 function. As might be expected from the relatively small difference in binding constant, MeCP2-induced interactions occurred with both methylated and unmethylated substrates.
In the present communication, we clarify the specific interactions between MeCP2 and fully defined DNA and chromatin substrates based on a systematic study of the complexes formed with wild-type MeCP2 and key RTT-inducing mutations. The source of DNA is a well-characterized 208-bp sequence with a strong nucleosome positioning sequence (40, 52) and 12 CpG sites (Fig. 1B). NAs are reconstituted on tandem repeats of the 208-bp DNA. This allows an experimental approach in which MeCP2 can be challenged with methylated or unmethylated arrays in the presence of excess unmethylated competitor having a different array length. The system also allows conformational changes induced by MeCP2 to be observed directly by electron microscopy (20). Using this strategy, we show that the binding of MeCP2 to DNA differs substantially from binding to chromatin reconstituted on the same sequence. MeCP2 binding results in significant conformational effects, and striking differences are seen with some MeCP2 mutants, providing important insights into the roles of the domains and the molecular mechanisms involved.
MATERIALS AND METHODS
Preparation of wild-type and mutant hMeCP2.
Plasmids containing genes for wild-type MeCP2 and the mutants R106W, R133C, and E397K were kindly supplied by Paul Wade and Timur Yusufzai. The additional constructs used in the present study, R294X, H370X, and R453X and the double mutants R106W/R294X and R106W/H370X, were engineered as described below.
Carboxy-terminal deleted coding sequences of MeCP2 with the 5′ ∼100-bp extension into the pTYB1 vector sequence bearing a NdeI site overlapping in frame with the MeCP2 start codon and a 3′ EcoRI linker (GACCGTGAATTC) were PCR amplified from full-length pTYB1-MeCP2 cDNA using the following primer pairs: R294X, Forward-5′-CCGGTTTAAACCGGGGATCTCGATCC-3′ and Reverse-5′-GACCGTGAATTCTCGGATAGAAGACTCCTTCACG-3′; H370X, Forward-5′-CCGGTTTAAACCGGGGATCTCGATCC-3′ and Reverse-5′-GTTAGAGAATTCGTGATGGTGGTGGTGCTCCTTCTTG-3′; and R453X, Forward-5′-CCGGTTTAAACCGGGGATCTCGATCC-3′ and Reverse-5′-GTTAGAGAATTCGTGTTTGTACTTTTCTGCGGCCGTGG-3′. The forward primer used in the amplification reactions was complementary to a site ∼100 bp upstream of the MeCP2 start codon in the pTYB1 vector.
The restriction fragments (NdeI + EcoRI) of the amplicons were then cloned into pTYB1 (New England Biolabs) vector by standard ligation procedures. The double mutants were constructed according to a similar procedure in which pTYB1-R106W was used as the template in the amplification reaction.
MeCP2 purification.
hMeCP2 and mutations were prepared as described previously (20) with modifications. The primary step in purification was performed by IMPACT (for intein-mediated purification with an affinity chitin-binding tag; New England Biolabs). After elution from the chitin agarose columns, the pooled MeCP2-containing fractions were freed of contaminating nucleic acids using a HiTrap heparin HP column (Amersham Biosciences) preequilibrated in 25 mM Tris-HCl, 5% glycerol, 250 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM benzamidine, and 30 μg of TPCK (tosyl phenylalanyl chloromethyl ketone)/ml. MeCP2 was applied to the column in 250 mM NaCl and eluted using salt steps from 0.2 to 1.0 M NaCl, increasing in increments of 0.1 M. The protein eluted at 800 mM salt. All preparations were checked for purity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and shown to consist of a single band; after storage, minor degradation (5 to 10%) was often observed.
Preparation of DNA and NAs.
Plasmids containing [208]12 DNA, kindly supplied by S. A. Grigoryev, were transformed into Stbl2 competent cells (Invitrogen). Transformants were propagated in Terrific Broth (Difco Corp), and plasmids were isolated by using Maxi kits (QIAGEN) according to the manufacturer's instructions.
[208]12 DNA was released by HhaI digestion, purified through a Sephacryl-1000 size exclusion column (Amersham), and recovered by isopropanol precipitation. DNA methylation was carried out using SssI methylase and verified by AvaI (DNA methylation-sensitive) digestion. [208]1 DNA was obtained by digestion of [208]12 DNA by EcoRI, followed by purification and ethanol precipitation. All restriction enzymes were obtained from New England Biolabs.
[208]12 NAs and [208]1 nucleosomes were prepared by mixing purified chicken erythrocyte core histones as described previously (40, 54) except that gradient dialysis from 2.0 M NaCl to 250 mM NaCl was used for array reconstitution, followed by exhaustive dialysis to 5 mM NaCl. NAs were checked on DNP gels for a single major band, and the mean number of nucleosomes per array determined by electron microscopy (EM).
Electrophoretic mobility shift assays (EMSAs).
For DNA, unmethylated or methylated target DNA (50 to 100 ng) was mixed with unmethylated competitor DNA (150 to 200 ng), followed by incubation with various amounts of MeCP2 in binding buffer (100 mM NaCl, 20 mM HEPES [or 10 mM Tris], 0.05% NP-40, 0.25 mM EDTA [pH 7.4]) at room temperature for 30 min.
Samples in which the target was [208]1 DNA were immediately electrophoresed on prechilled 6% polyacrylamide gel (mono/bis ratio of 35:1) in 0.5× TB (45 mM Tris, 45 mM borate [pH 8.3]) at 200 V for 4 h in cold conditions. For [208]12 DNA targets, electrophoresis was performed on prechilled 1% agarose type IV gels, which were run at 85 V for 4 h at 4C in TAE (40 mM Tris, 24 mM acetic acid, 0.5 mM EDTA [pH 8.3]) buffer.
For NAs, 200 ng of methylated or unmethylated target chromatin was mixed with 400 ng of unmethylated mononucleosome competitor in NA binding buffer (30 mM NaCl, 5 mM HEPES [pH 8.0], 0.5 mM EDTA, 0.025% NP-40), and MeCP2 was added at input ratios (based on target NA only) from one to four MeCP2 per [208]1 unit, followed by incubation for 30 min at room temperature. Electrophoresis was performed in 1% agarose (Sigma type IV) for 3.5 h at 85 V with TAE running buffer. DNA size markers were provided by the 1-kb Plus Ladder (Gibco-BRL).
DNA binding assay.
The volume of swollen CM-Sephadex beads (cat C25120; Sigma-Aldrich) sufficient to fully adsorb the input of MeCP2 in a DNA EMSA experiment was determined empirically. Methylated [208]12 DNA was incubated with wild-type or mutant MeCP2 at r = 2 or 3 under standard conditions; CM-Sephadex was added, followed by incubation for 40 min and then centrifugation to remove the Sephadex. The supernatants containing unbound DNA were analyzed on 1% agarose as described above.
DNA and chromatin gels were stained by ethidium bromide and photographed with the Kodak Gel 200 system, and band signals were quantitated by using Gel-Doc (Kodak). Gel shifts were quantitated by determining the electrophoretic mobility of the bands, and the effects of MeCP2 additions were measured by subtracting the mobilities of controls with no MeCP2.
Chromatin binding assay.
After the incubation of [208]12 NAs with MeCP2 under standard conditions, MgCl2 was added to 3.5 mM to promote the formation of NA oligomers, and the samples were centrifuged. This procedure resulted in the pelleting of NAs both in the absence and in the presence of MeCp2, but left unbound MeCP2 in the supernatants. Pellets and supernatants were mixed with SDS sample buffer, and the proteins were displayed by using SDS-16% PAGE.
NA oligomerization assay.
Target [208]12 NAs were incubated in NA binding buffer with a twofold amount of competitor mononucleosomes and an input MeCP2 ratio of four molecules per [208]1 unit of target NAs for 30 min at room temperature. The desired amount of MgCl2 was then added, and the samples were held for 20 min on ice and then centrifuged 10 min at 16,000 × g. The supernatants and pellets were separated and then treated with 1% SDS and proteinase K (0.2 mg/ml) for 1 h at 55C, and the products were separated on 1% agarose DNA gels.
Electron microscopy.
DNA and NA samples in 5 mM HEPES (pH 8.0), 0.5 mM EDTA, and the desired amounts of NaCl were placed in minidialyzer units (Pierce) and dialyzed for 4 h at 4C against buffer containing fresh 0.1% EM grade glutaraldehyde, followed by overnight dialysis against buffer alone. Samples were prepared for EM essentially as described previously (61). Briefly, material was adjusted to 50 mM NaCl, applied to glow-discharged carbon-coated 400-mesh grids, and rinsed three times with 5 mM Mg acetate (30) and 2% uranyl acetate used as a negative stain, or, after rinsing with water, as a positive stain. Some positively stained samples were subsequently lightly shadowed with Pt at an angle of 10°. In addition, some samples were prepared by spraying from glycerol, followed by shadowing with Pt (20). Positively stained and shadowed specimens were examined using tilted-beam dark-field imaging.
Grids were examined in a Tecnai 12 electron microscope (FEI Corp.) at 100 kV using a LaB6 filament, and images were recorded on a charge-coupled device camera (2048 × 2048; Tietz GMBH, Gauting, Germany). Image processing and measurement used the EMAN software package (39) or ImageJ.
The overall morphology of wild-type MeCP2 was calculated from a pool of 212 low-dose (∼5e/A2) particle images. Using EMAN single-particle reconstruction software, the images were grouped into self-similar classes by reference-free classification. The images in each class were aligned translationally and rotationally, and class averages, each representing a possible two-dimensional projection of the protein, were computed. Upon examination of the eight class averages generated, one was comprised of 89 of the original particles, whereas the other classes each contained 10 or fewer particles.
RESULTS
The basic substrate used for these studies is the 208-bp DNA sequence from the Lytechinus variegatus 5S RNA gene (51) that contains a strong nucleosomal positioning region (Fig. 1B). The sequence also has 12 CpG units, which, when methylated, become potential MeCP2 binding sites, and at least half meet the requirement for nearby A/T runs needed for maximal binding of MeCP2 (33). We used individual units or tandemly ligated arrays of 12 208-bp units (designated [208]1 and [208]12, respectively), either as naked DNA or as NAs prepared by reconstitution with purified core histones (see Materials and Methods for details). The biochemical, biophysical, and structural properties of [208]12 NAs have been extensively studied and reviewed (14, 25). To provide methylated substrates, DNA was treated with SssI prior to reconstitution, and digestion by AvaI was routinely used to verify complete methylation. No differences in reconstitution efficiency or products between methylated and nonmethylated substrates were observed (not shown). Recombinant human MeCP2 (splice variant e2 [8, 42]) and selected RTT-causing mutants (Fig. 1A) were expressed in Escherichia coli.
To monitor the interactions between MeCP2 and substrates, EMSAs were selected, with 6% acrylamide gels for [208]1 DNA and 1% agarose gels for [208]12 DNA and all experiments with NAs. This method was chosen in preference to others such as Southwestern blots (11) because the reactions occur in solution, and the products of any interactions can be analyzed both biochemically and by direct EM imaging.
Conditions for promoting methylation specific binding.
When DNA or NAs are exposed to wild-type MeCP2, complexes are formed that show a marked retardation when resolved on acrylamide or agarose gels, a property independent of the methylation status of the substrate (20). However, if excess competitor in the form of unmethylated [208] DNA or NAs are included in the reaction, methylation-specific retardation is consistently observed (Fig. 2 to 4), and our standard incubation conditions included a two- to fourfold excess of unmethylated competitor (see Materials and Methods for details). All MeCP2/DNA ratios (rMeCP2) are given in terms of the molar ratio of MeCP2 to total 208-bp units of target DNA or NAs. For [208]1 DNA targets the competitor was [208]12 DNA, and for [208]12 DNA the competitor was [208[1 DNA, while for [208]12 NAs the competitor was [208]1 mononucleosomes.
FIG. 2.
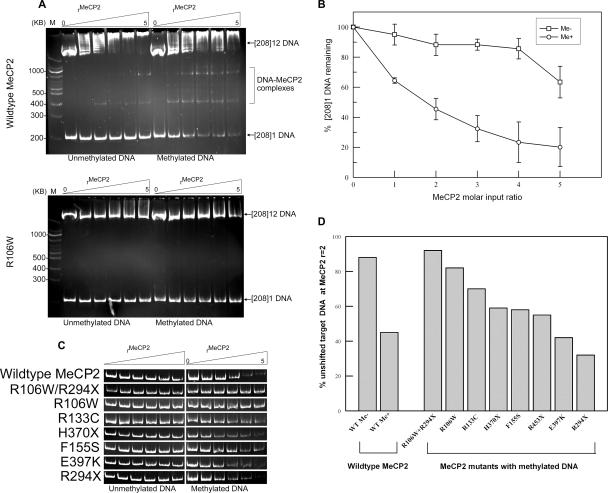
MeCP2 interacts with methylated [208]1 DNA. (A) Wild-type MeCP2 (upper gel) was incubated with unmethylated or methylated [208]1 DNA in the presence of a threefold excess of [208]12 DNA competitor at molar input ratios of zero to five MeCP2 per total [208] unit, and the products were displayed on 6% acrylamide gels. With methylated target DNA, there is a steady loss of the [208]1 DNA band and appearance of MeCP2-DNA complexes. In contrast, the MBD mutant R106W (lower gel) shows virtually no interaction with methylated or unmethylated DNA. (B) Quantitation of wild-type EMSA data from panel A. Each point is the mean and standard error of three or more experiments. (C) Comparison of the strength of the [208]1 DNA band for the MeCP2 mutants and constructs used in the present study (Fig. 1A). (D) Bar chart showing the percentage of unshifted target [208]1 DNA for MeCP2 mutants and constructs at MeCP2r = 2.
FIG. 4.
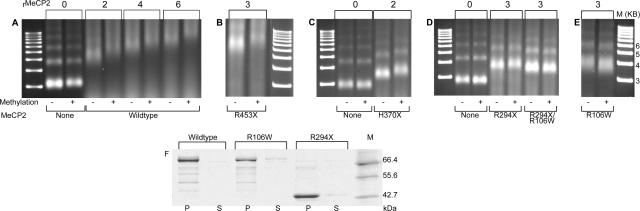
Interactions of wild-type MeCP2 and selected mutants and C-terminal truncations with [208]12 NAs. (A) Wild-type MeCP2 was incubated with unmethylated (−) or methylated (+) [208]12 NAs in the presence of a twofold excess of [208]1 mononucleosomes as competitor at molar input ratios of 0, 2, 4, and 6, and the products were displayed on 1% agarose gels. As with the [208]12 DNA target, there is a clear methylation-independent gel shift and a methylation-dependent enhancement. (B) The R453X truncation is very similar to wild-type MeCP2. (C) In contrast, the H370X truncation shows a pronounced diminution of the methylation-independent shift, and only a small methylation-induced enhancement. (D and E) R294X, R106W, and the double mutant R294X/R106W show only weak methylation-independent gel shifts which are unchanged with methylated substrates. (F) MeCP2 and the R106W and R294X mutants bind quantitatively to NAs. Methylated NAs were incubated with MeCP2 under standard conditions, MgCl2 was added to 3 mM, and the samples were pelleted. In all cases, the pelleted NAs contain at least 95% of input MeCP2.
Binding of MeCP2 to DNA.
Incubation of methylated [208]1 DNA with increasing amounts of wild-type MeCP2 (rMeCP2 = 0 to 5) results in a progressive reduction in the amount of unbound [208]1 DNA, and the appearance of discrete shifted bands (Fig. 2A, upper panel). In contrast, when the target is unmethylated, the interaction is much weaker. An estimate of the binding constant for wild-type MeCP2 to methylated DNA under these conditions is 4.5 × 10−8 M, very similar to the 4.0 × 10−8 M value obtained by Ballestar et al. (2). The various mutant forms of MeCP2 show striking differences in ability to induce gel shifts with the methylated [208]1 DNA substrate. The results for the R106W mutant shown in Fig. 2A (lower panel) illustrates these differences. This mutation shows no appreciable interaction with the methylated [208]1 target DNA. Further, while wild-type MeCP2 interacts with the unmethylated [208]12 competitor DNA (Fig. 2A, upper panel), this also does not occur with R106W, underscoring the remarkable loss of activity of this mutant. Figure 2C provides a comparison of the interactions of a variety of mutants with unmethylated and methylated [208]1 target DNA. A plot of the amount of unshifted [208]1 DNA versus wild-type MeCP2 input (Fig. 2B) indicates that ca. 50% of target DNA remains at rMeCP2 = 2.0, and the values for all mutants at this input ratio are summarized in the bar chart in Fig. 2D. The MBD mutant R106W gives a very weak interaction, comparable to the result with wild-type MeCP2 and unmethylated DNA, while interactions with E397K and the truncation mutant R294X are very similar to the wild type. For reasons that will become apparent below, we created the additional truncation mutants H370X and R453X and the double mutants R106W+R294X and R106W+H370X. None of the C-terminal truncation mutants differ substantially from the wild type, suggesting that this region has no role in methylation-dependent interactions with naked DNA, so that its complete or partial removal has no effect. In contrast, the double mutant R106W/R294X shows almost no interaction with methylated [208]1 DNA, underscoring the importance of this amino acid change within the MBD. Direct EM visualization suggests that the discrete slower-migrating bands that are especially prominent in methylation-dependent interactions (Fig. 2A, upper panel) are the result of the binding of one or more MeCP2 molecules to methylated sites (see below).
Interestingly, the unmethylated [208]12 competitor DNA which runs near the top of the gels (Fig. 2A) interacts differently with the various MeCP2 mutations. With wild type, the competitor itself becomes increasingly shifted as the MeCP2 input ratio is raised (Fig. 2A, upper panel), indicating an inherent methylation-independent interaction. This effect is, however, almost completely abolished with the R106W mutant (Fig. 2A, lower panel).
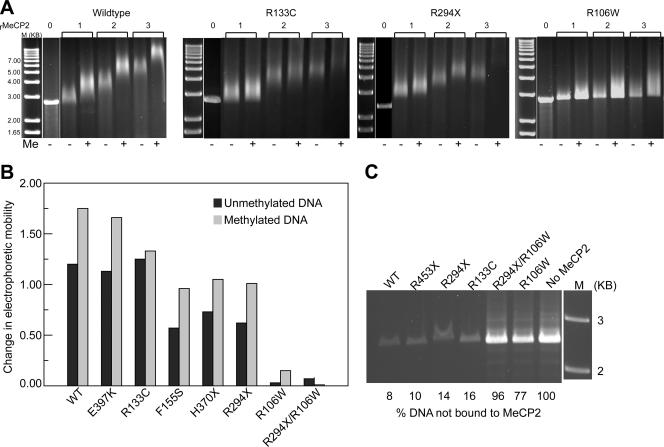
In terms of methylation-dependent EMSA effects, results similar to those obtained with [208]1 DNA are seen when the competitor and target substrates are reversed (Fig. 3). However, there are significant differences between the two data sets. First, instead of a gradual loss of unshifted target and the appearance of discrete intermediates, the [208]12 DNA substrates are shifted en masse at all MeCP2 input levels (Fig. 3A). This is probably due to the numerous (n = 144) methylated sites in these substrates, and consequently the large number of potential DNA-MeCP2 species, which will tend to form a diffuse band upon electrophoresis. Second, with [208]12 DNA, the unmethylated substrates typically show a reproducible gel shift (Fig. 3A and B) that is not competed out under our conditions. Examination of Fig. 3B shows that the interactions of MeCP2 with unmethylated substrates fall into three groups: the first, comprising E397K and R133C, shows the same mobility as the wild type. Significantly, R133C tends to give less severe RTT symptoms (see, for example, references 31, 32, 37, 53, and 62), perhaps as a result of the normal level of methylation-independent interactions. The second group, which includes F155S, H370X, and R294X, results in substantially weaker methylation-independent gel shifts. It is likely that for the truncation mutants, the reduced mass may account for the weak EMSA effect. The third group, containing R106W and the combination R106W/R294X, shows no interactions with either methylated or unmethylated substrates. Substantial methylation-dependent gel shift enhancement is seen with all of the mutants, with the exception of R133C, which shows only a small methylation-dependent shift, and R106W, which shows none. R133C is predicted to be deficient in binding methylated substrates (46).
FIG. 3.
MeCP2 interactions with target [208]12 DNA in the presence of competitor [208]1 DNA. (A) MeCP2 (wild type, R133C, R294X, and R106W) was incubated with unmethylated (−) or methylated (+) [208]12 DNA in the presence of [208]1 DNA competitor at molar input ratios of 0 to 3, and the products are displayed on 1% agarose gels. A methylation-dependent gel shift enhancement is seen with wild-type and R294X but not with R133C. The R106W mutant shows almost no methylation independent or dependent interactions. A narrow white space between lanes here and in panel C denotes places where irrelevant lanes have been removed from the image. (B) Bar chart summarizing results with MeCP2 mutants and constructs at r = 2. (C) Binding of MeCP2 (r = 2) to methylated [208]12 DNA. Shown is a 1% agarose gel of DNA remaining unbound after reacting with wild-type and mutant MeCP2. The R106W mutation results in a dramatic reduction in DNA binding.
We next determined the extent to which these EMSA changes result from differences in MeCP2-DNA binding using the observation that MeCP2 and DNA-MeCP2 complexes, but not free DNA, are strongly adsorbed to CM Sephadex beads. Methylated [208]12 DNA was incubated with MeCP2 at r = 2 and 3, sufficient Sephadex was added to adsorb all of the MeCP2, and the samples were centrifuged. Figure 3C shows the DNA content of supernatants from an experiment with MeCP2r = 3. As expected, the control (no MeCP2) shows the input level of [208]12 DNA, and with wild-type MeCP2 almost all of the DNA is bound with protein. Complete DNA binding was also observed with R133C, R294X, and R453X. The R106W mutant (and the double mutant with R294X) stand out as having a much-reduced capacity to bind DNA. Quantitation of the bands indicates that R106W has only ∼30% of the DNA-binding capacity of wild-type MeCP2. This emphasizes the remarkably powerful influence on DNA binding of the single amino acid change from arginine to tryptophan at this position. Further, since the loss of one arginine will have a negligible effect on the net positive charge, it is clear that net charge must play a rather minor role in MeCP2-DNA interactions.
Binding of MeCP2 to NAs.
Having characterized the interactions between MeCP2 and DNA, we sought to focus on a system that more closely approximates the in vivo chromatin substrate. Therefore, nucleosomes were assembled on methylated and unmethylated [208]1 and [208]12 DNA substrates by salt dialysis. The resulting NAs migrate as discrete bands on agarose gels (Fig. 4A, leftmost lanes). The minor bands that appear to be higher oligomers are not the result of contamination with longer DNA since DNA gels show a single species. Rather, direct EM observation shows that they result from occasional end-to-end self-association of NAs. The results of incubating methylated and unmethylated NAs with wild-type MeCP2 in the presence of unmethylated competitor [208]1 mononucleosomes are shown in Fig. 4A. As with [208]12 DNA, [208]12 NAs show a progressive methylation-independent gel shift and a consistent methylation-dependent shift enhancement, and the minor truncation mutant R453X gives a similar result (Fig. 4B). However, the more extensive C-terminal truncations yield much-reduced gel shifts. H370X shows a much weaker interaction with unmethylated NAs, and a slight enhancement with the methylated substrate (Fig. 4C), whereas R294X results in a weak methylation-independent effect and no methylation enhancement (Fig. 4D). This is in contrast to the situation with [208]12 DNA where the C-terminal truncations are as potent as wild-type MeCP2 in inducing methylation-dependent gel shift enhancement (Fig. 3) and suggests that the C terminus of MeCP2 harbors a domain (or domains) required for binding to chromatin. In its absence, both methylation-dependent and independent interactions are compromised, including the interaction between DNA and the MBD. Georgel et al. (20) reported that a R168X mutant also showed only a weak interaction with NAs. Further underscoring the importance of the C-terminal region of MeCP2, Chandler et al. (11) demonstrated reduced interactions between Xenopus laevis MeCP2 lacking part of the C terminus and mononucleosomes prepared from a 219-bp segment of the β-phaseolin promoter.
In keeping with the observed weak interaction between R106W and DNA (Fig. 3), this mutant is also ineffective when incubated with NAs (Fig. 4E), and the double R294X/R106W mutant (Fig. 4D) has the weakest methylation-independent gel shift, with no methylation enhancement. The only naturally occurring mutant showing a methylation-dependent gel shift is E397K (not shown). This also showed normal interactions with DNA (Fig. 2, 3), suggesting that its mild pathology is related to interactions with components other than chromatin.
Interpretation of EMSA data.
Although the EMSA approach provides a good indication of the magnitude of the interactions between components, it is not able to discriminate between the various mechanisms that could lead to a gel shift. In these experiments, shifts could be caused by one or more of the following: an increase in the mass of the complex (influenced by MeCP2 truncations), a change in the net charge, or a change in conformation (compaction and oligomerization). Conversely, the absence of a gel shift for a given mutant could be caused simply by a lack of binding. To address the latter possibility, we used the finding that NAs oligomerize and can be quantitatively pelleted in 3.5 mM MgCl2 (47), whereas free MeCP2 remains in solution. Wild-type MeCP2 and the R106W and R294X mutants were incubated with methylated NAs and, after the standard incubation, MgCl2 was added, the samples were centrifuged, and pellets and supernatants were separated by SDS-PAGE (Fig. 4F). In all three cases, the vast majority of the MeCP2 was in the pellet, indicating that the absence of or reduction in gel shifts seen with these mutants (Fig. 4D and E) is not due to the failure of MeCP2 to bind chromatin.
To help elucidate the nature of the interactions that result in the observed EMSA changes, we used direct EM visualization of the structural and/or conformational changes that accompany MeCP2 binding. Our strategy of using different array sizes for target and competitor provides an unambiguous way to differentiate between the methylated and unmethylated components.
MeCP2 is an oblate ellipsoid and binds cooperatively to methylated DNA.
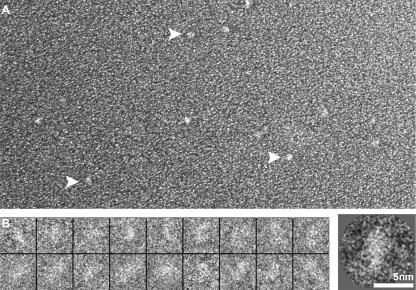
We first sought to characterize the shape of MeCP2 itself. Sizing columns and sedimentation values both indicate a much higher molecular mass than the 52 kDa predicted from the amino acid sequence, and yet there is no evidence for the formation of multimeric complexes (33). This suggests that MeCP2 has an elongated structure, and indeed, Klose and Bird (33) concluded from hydrodynamic measurements that MeCP2 was elongated, with a Stokes radius of ∼6 nm and a frictional coefficient f/f0 of ∼2.4. When observed in negative-stain EM (Fig. 5), wild-type MeCP2 appears as an oblate ellipsoid, and image averaging yields dimensions of ∼5.5 nm by ∼2.6 nm, indicating an axial ratio of 1:2.1.
FIG. 5.
MeCP2 is an oblate ellipsoid. (A) Field of wild-type hMeCP2 molecules shadowed with Pt. Individual uniformly sized particles are seen (arrows). (B) Gallery of negatively stained examples of wild-type MECP2 and, on the right, a two-dimensional average based on a pool of images.
Conformation of DNA-MeCP2 complexes.
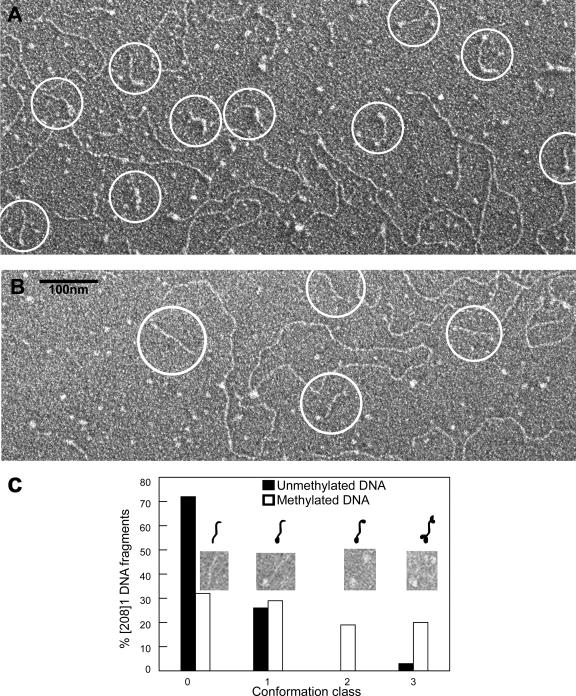
Electron micrographs of complexes formed between [208]1 DNA and wild-type MeCP2 in the presence of unmethylated [208]12 DNA as competitor are shown in Fig. 6A. Examples of the interactions with methylated and unmethylated DNA substrates are presented in the upper and lower panels of this figure, respectively. EMSA analysis of these interactions (Fig. 2A) reveals three principal components: unbound [208]1 DNA, unbound [208]12 competitor DNA, and DNA-MeCP2 complexes, seen as discrete shifted bands. EM images of this material reveal the short target DNA strands (circled in Fig. 6A), clearly distinct from the long competitor DNA. In the methylated sample, the [208]1 DNA strands are frequently decorated by one or more particles, which we interpret as bound MeCP2 molecules. In contrast, there are very few DNA strands with bound MeCP2 in the sample containing unmethylated target DNA. Analysis of the distribution of the different types of [208]1 DNA (Fig. 6B) suggests that the fastest-migrating intermediate band seen by EMSA corresponds to [208]1 DNA with one bound MeCP2 and that additional more slowly migrating bands correspond to DNA with two or more bound molecules.
FIG. 6.
Visualization of MeCP2 interactions with methylated and unmethylated [208]1 DNA. (A) Wild-type MeCP2 was incubated with target DNA in the presence of [208]12 competitor DNA at r = 4 (the same conditions as for Fig. 2A). With methylated DNA, many of the [208]1 DNA strands (circled) are decorated with particles, whereas most of the unmethylated DNA strands are undecorated (B). Long DNA strands are unmethylated [208]12 competitor DNA. (C) Bar chart comparing the frequency of four different classes of DNA-MeCP2 complexes with methylated and unmethylated target DNA. CpG methylation strongly promotes the binding of MeCP2 to DNA.
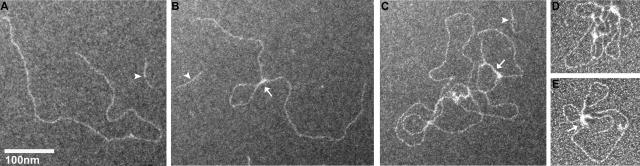
We next examined the reverse interaction in which the target DNA is methylated [208]12 DNA and the competitor is unmethylated [208]1 DNA, conditions producing the EMSA data seen in Fig. 3. Observations of the complexes formed with wild-type MeCP2 and the R106W and R294X mutants revealed two classes of interaction. With R106W, the material is indistinguishable from [208]12 DNA alone, showing no bound MeCP2 and no conformational changes (Fig. 7A). The second class of images, seen with both wild-type and R294X MeCP2, is characterized by striking conformational changes (Fig. 7B to E). The simplest conformation is a single loop resulting from an interaction within one target DNA molecule (Fig. 7B). Also common are more elaborate complexes containing two or more target DNA molecules characterized by extended regions in which two DNA segments are in close juxtaposition (Fig. 7C, arrow). These segments have the dimensions predicted for a DNA-MeCP2-DNA sandwich with a single ∼2 nm wide MeCP2 molecule between the two DNA molecules. Similar MeCP2-DNA conformations have been previously reported (20). In some instances, more complex structures are formed from which four or more DNA molecules emerge (Fig. 7D and E). MeCP2 shows a strong affinity for four-way junction DNA (19, 58), and analogous DNA conformations may occur within these complexes.
FIG. 7.
MeCP2 interacts with methylated DNA forming linear DNA-MeCP2-DNA complexes and loops. (A) A positive-stain EM image of the interaction of R106W MeCP2 with methylated [208]12 DNA at r = 3 shows no conformational changes, a finding consistent with the EMSA data (Fig. 3). The short DNA strand (arrowhead) is [208]1 competitor DNA. (B to E) In contrast, when reacted with the R294X truncation mutant (or wild type [data not shown]), dramatic conformational changes are seen. The DNA-MeCP2-DNA complexes include simple juxtapositions in cis (B, arrow), elaborate multistrand events (C), and loops (D and E). The arrow in panel C points to a region where two DNA strands have been brought together by MeCP2. Panels A to C are positively stained; panels D and E show Pt shadowed preparations.
In summary, the micrographs suggest that the initial interaction between MeCP2 (wild type and R294X) and DNA result in binding but no local DNA bending, which can subsequently spread cooperatively to form side-by-side interactions consisting of DNA-MeCP2-DNA sandwiches, and complex multistrand complexes. These properties may well contribute to the formation in vivo of MeCP2-dependent loops which appear to play important functional roles (26).
MeCP2 induces specific conformational changes in nucleosomal arrays.
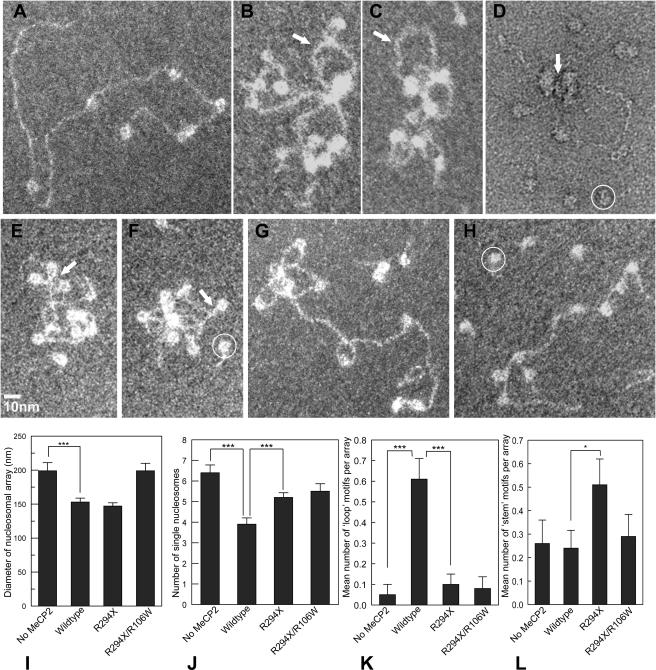
The EMSA data (Fig. 4) suggest that the binding of wild-type MeCP2 to nucleosomal arrays occurs in at least two distinct steps: (i) a methylation-dependent DNA-protein interaction requiring an intact MBD and methylated DNA and (ii) a methylation-independent interaction between the MeCP2 C-terminal domain and nucleosomes. To investigate the potential conformational changes associated with MeCP2 binding, we used direct EM imaging of the complexes formed between methylated NAs and wild-type MeCP2, as well as the R294X and R106W mutants. The analysis of NAs fully saturated with nucleosomes requires three-dimensional reconstructions of the compacted arrays (20) and, even then, the locations of linker DNA are often obscured. In contrast, subsaturated NAs show minimal intrinsic folding (14), and the regions of free DNA interspersed with nucleosomes allow better discrimination between binding to DNA and binding to nucleosomes. Therefore, we created subsaturated [208]12 NAs containing on average seven nucleosomes and observed the conformational changes induced by MeCP2 and selected mutants. Although the subsaturated arrays alone show no compaction (Fig. 8A), profound conformational changes occur in the presence of wild-type MeCP2, with clusters of nucleosomes from which loops of free DNA emanate (Fig. 8B to D). This supports the concept that array compaction contributes significantly to the observed gel shifts (Fig. 4). In negatively stained preparations, it was sometimes possible to see MeCP2-sized material sandwiched between nucleosomes (Fig. 8D). Importantly, the R294X mutant, which interacts strongly with DNA but not nucleosomes, induced some compaction, but via a different type of conformational change. With this mutant, the most prominent motif is caused by DNA-DNA juxtapositioning, which is especially prominent close to the nucleosomes, bringing the entering and exiting linker DNA close together (Fig. 8E and F), thus creating a “stem” motif from the paired linkers. As anticipated from the weak EMSA shift, the R106W mutant induces hardly any conformational changes (Fig. 8G), and the double mutant R106W/R294X is equally impotent (Fig. 8G).
FIG. 8.
Undersaturated [208]12 NAs provide useful insights into the types of conformational change induced in chromatin by MeCP2. (A) Typical undersaturated array with seven nucleosomes. (B and C) Methylated arrays after incubation with wild-type MeCP2 at r = 4 in the presence of [208]1 mononucleosomes as competitor (circled in panels D, F, and H). Significant compaction has occurred, with nucleosome clustering and DNA loop formation (arrows). (D) Same conditions as for panels B and C, but negative staining reveals a particle of the size and shape of MeCP2 sandwiched between nucleosomes (arrow). (E and F) With the R294X mutant, nucleosome clustering is minimal, but DNA-MeCP2-DNA interactions are common, especially near the linker entry-exit sites on nucleosomes, forming “stem” motifs (arrows). (G and H) In contrast, with the R106W mutant (G) or the double R106W/R294X (H) very few conformational changes are seen. (I to L) Bar charts comparing the influence of mutants on the various conformational changes. ***, P < 0.001; *, P = 0.05. (I) Compaction, as indicated by array diameter. (J) Nucleosome clustering, as indicated by the number of separate individual nucleosomes. (K) Frequency of loop motifs emanating from nucleosome clusters. (L) Frequency of stem motifs.
To provide a more quantitative indication of the different conformational changes, the complexes were scored for (i) overall compaction (measured by the diameter of the smallest circle enclosing the whole complex), (ii) the formation of close nucleosome-nucleosome contacts (measured by the number of separate individual nucleosomes resolved by EM in the array), (iii) binding to free DNA versus nucleosomes (measured by the presence of DNA loops emanating from a cluster of touching nucleosomes), and (iv) binding to the nucleosome linker entry site (measured by the number of “stem” motifs in individual nucleosomes). As expected from the EMSA data (Fig. 4), wild-type MeCP2 induces a highly significant array compaction, whereas compaction is essentially unchanged with the R106W mutant (Fig. 8I). With the R294X C-terminal truncation, compaction as measured by array diameter is similar to the wild type but, as noted above, the micrographs show this to be due primarily to DNA-DNA rather than to nucleosome-nucleosome interactions (Fig. 8F to H). Evidently, this type of interaction has only a minor effect on electrophoretic mobility (Fig. 4). As expected from the compaction measurements, the number of separate individual nucleosomes is significantly reduced with wild-type MeCP2 (due to nucleosome clustering) but not with R106W or R294X (Fig. 8J). The looping out of free DNA from nucleosome clusters, indicating nucleosome-nucleosome bridging (as opposed to DNA-DNA bridging), is also maximal with wild-type MeCP2, whereas the mutants R294X and R106W, like control untreated arrays, show very little looping (Fig. 8K). As expected, the frequency of the stem motif is greatly enhanced with the R294X mutant (Fig. 8L). With wild-type MeCP2, nucleosome clustering obscures many of the linker entry-exit sites and most likely accounts for the lack of the expected increase in the occurrences of stem motifs in this case.
MeCP2 induces oligomerization of NAs.
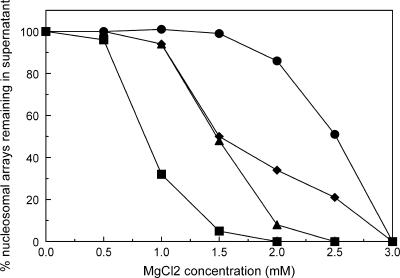
Our EMSA data show that the interaction between MeCP2 and NAs induces in a wide spectrum of mobility shifts, often including complexes too large to enter the 1% agarose gels. This phenomenon is correlated with the appearance of array oligomers in EM preparations (not shown), in agreement with the previous report (20). To determine whether key MeCP2 mutants had altered abilities to promote oligomerization, we used an assay based on the increased sensitivity of oligomeric arrays to precipitation by MgCl2 (47, 51). Saturated nucleosomal arrays were incubated under our standard conditions (including [208]1 competitor mononucleosomes) with wild-type and mutant MeCP2 at r = 4 and then exposed to a range of MgCl2 concentrations, followed by centrifugation. DNA was prepared from pellets and supernatants, separated on agarose gels, and the bands corresponding to [208]12 DNA were quantitated. The results (Fig. 9) show that ∼50% of the NAs are pelleted at a MgCl2 concentration of ∼3.0 mM, similar to previous reports (47, 51), whereas wild-type MeCP2 induces a much greater sensitivity to Mg2+, with 50% oligomerization at ∼0.75 mM. The mutants R294X and R106W both induced an intermediate level of Mg2+ sensitivity. These results further indicate that MeCP2 is a powerful promoter of array oligomerization, a property that is significantly reduced both in the R106W mutant and in C-terminal truncations.
FIG. 9.
MeCP2 influences array oligomerization. NAs were incubated with MeCP2 at r = 4, exposed to different amounts of MgCl2, and centrifuged. Without MeCP2 (•) 50% of the arrays oligomerize at ∼2.5 mM MgCl2, whereas with wild-type MeCP2 (▪) 50% oligomerization requires only ∼0.75 mM MgCl2. The mutants R106W (▴) and R294X (⧫) induce 50% oligomerization at ∼1.5 mM MgCl2.
DISCUSSION
This investigation was prompted by the unexpected finding that MeCP2, mutations of which cause at least 85% of RTT cases, induces dramatic compaction of chromatin (20). MeCP2 is thought to transmit epigenetic DNA methylation signals by promoting local transcriptional repression, and chromatin compaction is likely to contribute to this effect. Our goals were to understand the molecular mechanisms leading to compaction and to determine the effects on compaction of common RTT-inducing mutations. Also, since MeCP2 binds preferentially to methylated CpG sites on DNA, it was important to understand the relationship between methylation-dependent and -independent effects. Our strategy involves a combined approach using EMSAs and direct EM visualization. Interactions with recombinant human MeCP2 and key mutations were examined using three substrates, all based on a naturally occurring 208-bp DNA sequence containing a strong nucleosome positioning region (52). These can be methylated as desired, prepared as tandem 12-unit DNA arrays or reconstituted into NAs (Fig. 1B). To examine methylation dependence, excess unmethylated substrate differing in size from the methylated target was included in the reactions.
EMSA data using methylated [208]1 DNA as target and [208]12 DNA as competitor indicate a wide range of effects, depending on the type of MeCP2 (Fig. 2). The wild-type protein shows a clear methylation-dependent gel shift, a property shared with the E397K mutant and with all C-terminal truncations. At the other extreme was the MBD mutant R106W, which induces essentially no change in electrophoretic mobility of the DNA substrate. When the methylated target was [208]12 DNA, the same general pattern emerged, although there was a consistent methylation-independent shift with all except R106W (Fig. 3). The strikingly different effects observed with R106W and R294X prompted us to focus on these two naturally occurring MeCP2 mutations.
Since R294X behaves very similarly to wild-type in its interactions with DNA, we conclude that the C-terminal region beyond residue 294 is not needed for DNA binding. However, since this mutation produces RTT in humans, and a mouse strain engineered to lack residues beyond 308 exhibits many RTT-like symptoms, it is clear that the C-terminal region has a critical role in vivo not connected with binding to methylated DNA. R106W represents the opposite extreme, showing only very weak EMSA interactions with methylated DNA (Fig. 2 and 3). Although this mutation is within the MBD, nuclear magnetic resonance data suggest that it is not directly involved in DNA contacts (46), and its position is not significantly changed upon binding to DNA (60). Thus, there is no obvious reason why the arginine-to-tryptophan change should give rise to such a strongly altered phenotype. A unique aspect of R106W that contributes to its weak EMSA interactions is its weak binding to DNA (Fig. 3C). In contrast, the other mutants tested were all fully bound to [208]12 DNA under our interaction conditions (Fig. 3C). Since the loss of one arginine will have a negligible effect on the strong net positive charge of MeCP2, it is clear that even methylation-independent MeCP2-DNA binding involves more than simple electrostatic interactions.
EM examination of gel-shifted complexes formed between MeCP2 and methylated [208]12 DNA reveals profound conformational changes (Fig. 7), providing more insight into DNA-MeCP2 interactions. In agreement with the EMSA data, both wild-type and R294X induced a side-by-side juxtaposition of DNA, and the width of the sandwiched regions indicates that they are formed by DNA-MeCP2-DNA complexes containing a single file of MeCP2. This raises the possibility that MeCP2 has one or more DNA-binding regions in addition to the MBD with the additional DNA-binding site(s) occurring within residues 1 to 294.
Chromatin, which is closer to the in vivo MeCP2 substrate than naked DNA, reveals additional levels of interaction complexity. With NAs, methylation-dependent gel shift enhancement is seen principally with wild-type and the very minor R453X truncation (Fig. 4). R294X is again very informative, since, unlike the situation with DNA, it now shows no methylation effect, and its interaction with unmethylated chromatin is much weaker than wild-type (Fig. 4D). This indicates that the C-terminal region contains one or more chromatin-interacting regions and that, in their absence, the methylation-dependent MBD-based interaction is severely compromised. We can exclude the possibility that the weak interaction with R294X is due simply to low binding—under our conditions, the wild type and the R294X and R106W mutants are equally well bound to NAs (Fig. 4F). The H370X construct also gives a much weaker gel shift than does the wild type (Fig. 4C), but there is a small methylation-dependent effect. This suggests that interactions with chromatin are distributed over much of the C-terminal region.
Methylation-dependent and independent interactions.
The binding constant of MeCP2 to methylated DNA is ∼4 × 10−8 M (3; the present study) and differs by a factor of only 3 from nonspecific binding (15). Even with such a small difference in affinity, the differential binding of MeCP2 to methylated versus nonmethylated DNA is likely to be biologically relevant. The protein appears to have a level of mobility in vivo similar to that of histone H1 (41), with a mean residence half-life of ∼25 s (33), and differential binding will tend to increase the residence time on methylated DNA.
Other regulatory proteins which bind to specific sequence DNA also typically show nonspecific binding, a classic case being the lac repressor (18). In this instance, X-ray structures reveal that the molecular interaction between protein and DNA is quite different for specific and nonspecific binding (27), and it has been suggested that the nonspecific interaction plays an important biological function by dramatically reducing the time needed for the protein to encounter its specific binding sequence (21, 59). Similar arguments have recently been put forward for eukaryotic systems (23, 28, 29).
MeCP2 is a chromatin architectural protein.
The binding of proteins to chromatin can have a number of consequences. Even the simplest situation where the interaction results in no conformational change will effectively increase the mass of the chromatin and result in a gel shift. Additional phenomena that can alter gel shifts are charge neutralization, changes in conformation (compaction or expansion), and oligomerization. To distinguish between these phenomena, we used direct EM visualization of NAs after MeCP2 treatments that produced well-defined EMSA changes. As shown in Fig. 8 and reported previously (20), the predominant change in conformation resulting from MeCP2 binding to NAs is compaction. However, there are numerous routes that could lead to compaction, and to discriminate between them we made use of subsaturated NAs, which do not fold in response to increased salt (47). Subsaturated arrays also provide a much clearer distinction between binding to free DNA as opposed to binding to nucleosomes and allow quantitative comparisons of relative compaction (16).
When applied to wild-type MeCP2 and the R106W and R294X mutants, this strategy was very informative. The expected compaction induced by wild-type MeCP2 was clearly seen, whereas mutants with the R106W defect failed to compact (Fig. 8G and H). Surprisingly, the R294X truncation, which produces only a minimal gel shift, resulted in wild-type compaction, as measured by the diameter of the smallest circle encompassing the array (Fig. 8E and F). However, arrays compacted by wild-type and R294X MeCP2 were very different in conformation. While the wild-type protein tended to bring nucleosomes close together, this was rare with R294X. Rather, with R294X, individual nucleosomes remained separate, whereas DNA-DNA interactions effectively compacted the arrays (Fig. 8E and F).
Chromatin structural motifs induced by MeCP2.
Imaging subsaturated arrays also provides important insight into the types of interaction that lead to compaction. With wild-type MeCP2, the dominant motif is a loop, either of naked DNA or of DNA with one or more nucleosomes, emanating from a cluster of nucleosomes. This suggests that the dominant interaction involves nucleosome-nucleosome interactions (rather than DNA-DNA or DNA-nucleosome interactions). Indeed, it is possible to image particles of the dimensions of single MeCP2 molecules within nucleosome clusters (Fig. 8D). This observation, together with the report that MeCP2 does not form dimers or higher oligomers (33), suggests that single MeCP2 molecules are sandwiched between nucleosomes to create the motif. These nucleosome-MeCP2-nucleosome interactions require that MeCP2 has at least two chromatin-binding sites. The present data do not allow us to pinpoint these sites but do indicate that, within the C-terminal region, contributions from residues 294 to 370 and 371 to 453 are required for wild-type activity.
The second structural motif is the formation of a “stem” composed of linker DNA at the nucleosome entry-exit site, a conformation characteristic of H1-containing chromatin (5, 10, 24). This motif is most prominent in arrays compacted by the R294X mutant (Fig. 8E and F). It is also likely to be present in arrays compacted by wild-type MeCP2 but obscured by nucleosome-nucleosome interactions. DNA conformations similar to the linker entry-exit site also occur in four-way junction DNA, and it is notable that both MeCP2 and H1 bind strongly to four-way junctions (19, 58). This binding mode is MBD dependent but methylation independent (19). The R106W mutant gave only background levels of stem motifs.
Insights into molecular mechanisms.
A clear conclusion from the work with the R294X C-terminal mutant is that this domain is crucial for binding MeCP2 to chromatin. With DNA substrates, R294X is essentially identical to the wild type in methylation-specific binding but, despite the presence of a fully functional MBD, interacts only very weakly with chromatin (Fig. 4). This suggests that the binding of MeCP2 to chromatin occurs in two steps: a methylation-independent interaction between chromatin and the C terminus that is required for the second, methylation-specific, interaction between DNA and the MBD. Although the linker DNA in these arrays is quite long (∼50 bp) and contains three CpG dinucleotides (Fig. 1A), by itself it does not constitute a strong binding site in the context of chromatin and is unable to bind the R294X mutant in a methylation-dependent manner. This may account for the severity of RTT caused by this mutation, as well as the RTT-like phenotype of mice bearing MeCP2 lacking residues beyond 308 (49).
The MBD mutant R106W gave the most surprising results. Modeling based on nuclear magnetic resonance studies of the methyl binding domain of the related MBD1 protein suggests that this arginine does not directly interact with DNA (46, 60) and might be predicted to have little effect on methylation-dependent protein binding and induce only a low-severity phenotype. Instead, R106W completely abolishes methylation-specific binding both to DNA and chromatin and dramatically reduces methylation-independent binding to chromatin. Using Xenopus recombinant MeCP2, Ballestar et al. (2) also reported a dramatic (>100-fold) drop in the binding of R106W to a methylated DNA substrate compared to the wild type. One possible explanation of these observations is that the R106W mutation causes substantial changes in protein folding that lead to its aberrant behavior.
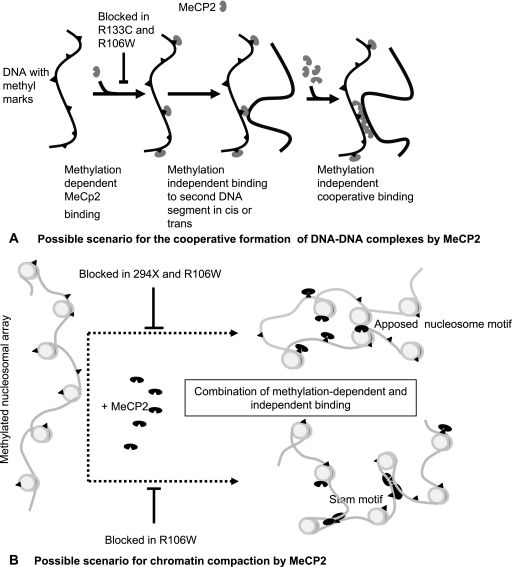
Possible scenarios for the major conformational changes observed in the present study are outlined in Fig. 10. Figure 10A illustrates the proposed interaction between MeCP2 and methylated DNA leading to the conformational changes shown in Fig. 6, and Fig. 10B suggests how the dual interactions between MeCP2 and nucleosomes may lead to chromatin compaction. It will clearly be important in future work to further define the chromatin component(s) and domain(s) of MeCP2 that are involved in the methylation-dependent and -independent interactions with nucleosomes.
FIG. 10.
Possible models for the induction by MeCP2 of conformational changes in DNA (A) and NAs (B).
An important conclusion highlighted by the comparison of the MeCP2 mutants is that the interactions between chromatin and MeCP2 are very different from the interactions with DNA. Further detailed studies using nucleosomal arrays will be needed to fully characterize the molecular events involved in these interactions and assess their potential importance in RTT.
Acknowledgments
We thank P. Wade and T. Yusufzai for hMeCP2 protein and plasmids.
This study was supported by the Rett Syndrome Research Foundation and NIH grants GM070897 to C.L.W. and GM45916 and GM66834 to J.C.H.
Footnotes
Published ahead of print on 13 November 2006.
REFERENCES
- 1.Amir, R. E., I. B. den Veyber, M. Wan, C. Q. Tran, U. Francke, and H. Y. Zoghbi. 1999. Rett syndrome is caused by mutations in X-linked MeCP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23:185-188. [DOI] [PubMed] [Google Scholar]
- 2.Ballestar, E., T. M. Yusufzai, and A. P. Wolffe. 2000. Effects of Rett syndrome mutations of the methyl-CpG binding domain of the transcriptional repressor MeCP2 on selectivity for association with methylated DNA. Biochemistry 39:7100-7106. [DOI] [PubMed] [Google Scholar]
- 3.Ballestar, E., S. Ropero, M. Alimanos, J. Armstrong, F. Setien, R. Agrelo, M. F. Fraga, M. Herranz, S. Avila, M. Pineda, E. Monros, and M. Esteller. 2005. The impact of MECP2 mutations in the expression pattern of Rett syndrome patients. Hum. Genet. 116:91-104. [DOI] [PubMed] [Google Scholar]
- 4.Bassal, K. H., C. Tikellis, and A. El-Osta. 2004. Expression analysis of the epigenetic methyltransferases and methyl-CpG binding protein families in the normal B-cell and chronic lymphocytic leukemia (CLL). Cancer Biol. Ther. 3:989-994. [DOI] [PubMed] [Google Scholar]
- 5.Bednar, J., R. A. Horowitz, S. A. Grigoryev, L. M. Carruthers, J. C. Hansen, A. B. Koster, and C. L. Woodcock. 1998. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher order folding and compaction of chromatin. Proc. Natl. Acad. Sci. USA 95:14173-14178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernard, D., J. Gil, P. Dumont, S. Rizzo, D. Monte, B. Quatannens, D. Hudson, T. Visakorpi, F. Fuks, and Y. de Launoit. 2006. The methyl-CpG-binding protein MeCP2 is required for prostate cancer cell growth. Oncogene 25:1358-1366. [DOI] [PubMed] [Google Scholar]
- 7.Bienvenu, T., and J. Chelly. 2006. Molecular genetics of Rett syndrome: when DNA methylation goes unrecognized. Nat. Rev. Genet. 7:415-426. [DOI] [PubMed] [Google Scholar]
- 8.Bird, A. P., and S. Kriaucionis. 2004. The major form of MeCP2 has a novel N terminus generated by alternative splicing. Nucleic Acids Res. 32:1818-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buschdorf, J. P., and W. H. Stratling. 2004. A WW domain binding region in methyl-CpG-binding protein MeCP2: impact on Rett syndrome. J. Mol. Med. 82:135-143. [DOI] [PubMed] [Google Scholar]
- 10.Carruthers, L. M., J. Bednar, C. L. Woodcock, and J. C. Hansen. 1998. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays. Biochemistry 37:14776-14787. [DOI] [PubMed] [Google Scholar]
- 11.Chandler, S. P., D. Guschin, N. Landsberger, and A. P. Wolffe. 1999. The methyl-CpG binding transcriptional repressor MeCP2 stably associates with nucleosomal DNA. Biochemistry 38:7008-7018. [DOI] [PubMed] [Google Scholar]
- 12.Chen, R. Z., S. Akbarian, M. Tudor, and R. Jaenisch. 2001. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat. Genet. 27:327-331. [DOI] [PubMed] [Google Scholar]
- 13.Fang, Y., J. Thiesen, and W. H. Stratling. 2000. Histone deacetylase-independent transcriptional repression by methyl-CpG-binding protein 2. Nucleic Acids Res. 28:2201-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fletcher, T. M., and J. C. Hansen. 1996. The nucleosomal array: structure/function relationships. Crit. Rev. Eukaryot. Gene Expr. 6:149-188. [DOI] [PubMed] [Google Scholar]
- 15.Fraga, M. F., E. Ballestar, G. Montoya, P. Taysavang, P. A. Wade, and M. Esteller. 2003. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 31:1765-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francis, N. J., R. E. Kingston, and C. L. Woodcock. 2004. Chromatin compaction by a Polycomb group protein complex. Science 306:1574-1577. [DOI] [PubMed] [Google Scholar]
- 17.Free, A., R. I. D. Wakefield, B. O. Smith, D. T. F. Dryden, P. N. Barlow, and A. P. Bird. 2001. DNA recognition by the methyl-CpG binding domain of MeCP2. J. Biol. Chem. 276:3353-3360. [DOI] [PubMed] [Google Scholar]
- 18.Fried, M., and D. M. Crothers. 1981. Equilibria and kinetics of the lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 9:6505-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galvao, T. C., and J. O. Thomas. 2005. Structure-specific binding to four-way junction DNA through its methyl CpG-binding domain. Nucleic Acids Res. 33:6603-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Georgel, P. T., R. A. Horowitz-Scherer, N. Adkins, C. L. Woodcock, P. A. Wade, and J. C. Hansen. 2003. Chromatin compaction by human MeCP2: assembly of novel secondary structures in the absence of DNA methylation. J. Biol. Chem. 278:32181-32188. [DOI] [PubMed] [Google Scholar]
- 21.Gowers, D. M., and S. E. Halford. 2003. Protein motion from nonspecific to specific DNA by three-dimensional routes aided by supercoiling. EMBO J. 22:1410-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guy, J., B. Hendrich, M. Holmes, J. E. Martin, and A. Bird. 2001. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat. Genet. 27:322-326. [DOI] [PubMed] [Google Scholar]
- 23.Halford, S. E., and J. F. Marko. 2004. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 32:3040-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamiche, A., P. Schultz, V. Ramakrishnan, P. Oudet, and A. Prunell. 1996. Linker histone-dependent DNA structure in linear mononucleosomes. J. Mol. Biol. 257:30-42. [DOI] [PubMed] [Google Scholar]
- 25.Hansen, J. C. 2002. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 31:361-392. [DOI] [PubMed] [Google Scholar]
- 26.Horike, S.-I., S. Cai, M. Miyano, J.-F. Cheng, and Kohwi-Shigematsu. 2005. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 37:31-40. [DOI] [PubMed] [Google Scholar]
- 27.Kalodimos, C. G., N. Biris, A. M. J. J. Bonvin, M. M. Levandoski, M. Guennueges, R. Boelens, and R. Kaptein. 2004. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science 305:386-389. [DOI] [PubMed] [Google Scholar]
- 28.Kampmann, M. 2004. Obstacle bypass in protein motion along DNA by two-dimensional rather than one-dimensional sliding. J. Biol. Chem. 279:38715-38720. [DOI] [PubMed] [Google Scholar]
- 29.Kampmann, M. 2005. Facilitated diffusion in chromatin lattices: mechanistic diversity and regulatory potential. Mol. Microbiol. 57:889-899. [DOI] [PubMed] [Google Scholar]
- 30.Katsani, K. R., M. A. N. Hajibagheri, and C. P. Verrijzer. 1999. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 18:698-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerr, A. M., and R. J. Prescott. 2005. Predictive value of the early clinical signs in Rett disorder. Brain Res. 27:(Suppl. 1)S20-S24. [DOI] [PubMed] [Google Scholar]
- 32.Kerr, A. M., H. L. Archer, J. C. Evans, R. J. Prescott, and F. Gibbon. 2006. People with MECP2 mutation-positive Rett disorder who converse. J. Intellect Disabil. Res. 50:(Pt. 5)386-394. [DOI] [PubMed] [Google Scholar]
- 33.Klose, R. J., and A. P. Bird. 2004. MeCP2 behaves as an elongated monomer that does not stably associate with the Sin3a chromatin remodeling complex. J. Biol. Chem. 279:46490-46496. [DOI] [PubMed] [Google Scholar]
- 34.Klose, R. J., S. A. Sarraf, L. Schmiedenberg, S. M. McDermott, I. Stancheva, and A. P. Bird. 2005. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol. Cell 19:667-678. [DOI] [PubMed] [Google Scholar]
- 35.Kudo, S. 1998. Methyl-CpG-binding protein MeCP2 represses Sp1-activated transcription of the human leukosialin gene when the promoter is methylated. Mol. Cell. Biol. 18:5492-5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LaSalle, J. M. 2004. Paradoxical role of methyl-CpG-binding protein 2 in Rett syndrome. Curr. Top. Dev. Biol. 59:61-86. [DOI] [PubMed] [Google Scholar]
- 37.Leonard, H., L. Colvin, J. Christodoulou, T. Schiavello, S. Williamson, M. Davis, D. Ravine, S. Fyfe, N. de Klerk, T. Matsuishi, I. Kondo, A. Clarke, S. Hackwell, and Y. Yamashita. 2003. Patients with the R133C mutation: is their phenotype different from patients with Rett syndrome with other mutations? J. Mol. Genet. 40:e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis, J. D., R. R. Meehan, W. J. Henzel, I. Maurer-Fogy, P. Jeppsen, F. Klein, and A. Bird. 1992. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69:905-914. [DOI] [PubMed] [Google Scholar]
- 39.Ludtke, S. J., P. R. Baldwin, and W. Chiu. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82-97. [DOI] [PubMed] [Google Scholar]
- 40.Meersseman, G., S. Penning, and E. M. Bradbury. 1991. Chromatosome positioning on assembled long chromatin: linker histones affect placement on 5S rDNA. J. Mol. Biol. 220:89-100. [DOI] [PubMed] [Google Scholar]
- 41.Misteli, T., A. Gunjan, R. Hock, M. Bustin, and D. T. Brown. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature 408:877-881. [DOI] [PubMed] [Google Scholar]
- 42.Mnatzakanian, G. N., H. Lohi, I. Munteanu, S. E. Alfred, T. Yamada, P. J. M. MacLeod, J. R. Jones, S. W. Scherer, N. C. Schanen, M. J. Friez, J. B. Vincent, and B. A. Minassian. 2004. A previously unidentified MeCP2 open reading frame defines a new protein isoform relevant to Rett syndrome. Nat. Genet. 36:339-341. [DOI] [PubMed] [Google Scholar]
- 43.Muller, H. M., H. Fiegl, G. Goebel, M. M. Hubalek, A. Widschwendter, E. Muller-Holzner, C. Marth, and M. Widschwendter. 2003. MeCP2 and MBD2 expression in human neoplastic and non-neoplastic breast tissue and its association with estrogen receptor status. Br. J. Cancer 89:1934-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nan, X., F. J. Campoy, and A. Bird. 1997. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 88:471-481. [DOI] [PubMed] [Google Scholar]
- 45.Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386-389. [DOI] [PubMed] [Google Scholar]
- 46.Ohki, I., N. Shimotake, N. Fujita, J.-G. Jee, T. Ikegami, M. Nakao, and M. Shirakawa. 2001. Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell 105:487-497. [DOI] [PubMed] [Google Scholar]
- 47.Schwarz, P. M., A. Felthauser, T. M. Fletcher, and J. C. Hansen. 1996. Reversible oligonucleosome self-association: dependence on divalent cations and core histone tail domains. Biochemistry 35:4009-4015. [DOI] [PubMed] [Google Scholar]
- 48.Shabazian, M. D., and H. Zoghbi. 2002. Rett syndrome and MeCP2: linking epigenetics and neuronal function. Am. J. Hum. Genet. 71:1259-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shabazian, M. D., J. I. Young, L. A. Yuva-Paylor, C. M. Spencer, B. A. Antalffy, J. L. Noebels, D. L. Armstrong, R. Paylor, and H. Zoghbi. 2002. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron 35:243-254. [DOI] [PubMed] [Google Scholar]
- 50.Sharma, D., J. Blum, X. Yang, N. Beaulieu, A. R. Macleod, and N. E. Davidson. 2005. Release of methyl CpG binding proteins and histone deacetylase 1 from the estrogen receptor alpha (ER) promoter upon reactivation in ER-negative human breast cancer cells. Mol. Endocrinol. 19:1714-1751. [DOI] [PubMed] [Google Scholar]
- 51.Shogren-Knaak, M., H. Ishii, M. J. J.-M. Sun Pazin, J. R. Davie, and C. L. Peterson. 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311:844-847. [DOI] [PubMed] [Google Scholar]
- 52.Simpson, R. T., and D. W. Stafford. 1983. Structural features of a phased nucleosome core particle. Proc. Natl. Acad. Sci. USA 80:51-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smeets, E., E. Schollen, U. Moog, G. Matthijs, J. Herbergs, S. Smeets, L. Curfs, C. Schrander-Stumpel, and J. P. Fryns. 2003. Rett syndrome in adolescent and adult females: clinical and molecular genetic findings. Am. J. Med. Genet. A 122:227-233. [DOI] [PubMed] [Google Scholar]
- 54.Springhetti, E. M., N. E. Istomina, J. C. Whisstock, T. Nikitina, C. L. Woodcock, and S. A. Grigoryev. 2003. Role of the M-loop and reactive center loop domains in the folding and bridging of nucleosome arrays by MENT. J. Biol. Chem. 278:43384-43393. [DOI] [PubMed] [Google Scholar]
- 55.Tada, Y., R. M. Brena, B. Hackanson, C. Morrison, G. A. Otterson, and C. Plass. 2006. Epigenetic modulation of tumor suppressor CCAAT/enhancer binding protein alpha activity in lung cancer. J. Natl. Cancer Inst. 98:396-406. [DOI] [PubMed] [Google Scholar]
- 56.Tryndyak, V. P., O. Kovalchuk, and I. P. Pogribny. 2006. Loss of DNA methylation and histone lysine 20 methylation in human breast cancer cells is associated with aberrant expression of DNA methyltransferase 1, SuV-20h2 histone methyltransferase and methyl-binding proteins. Cancer Biol. Ther. 5:65-70. [DOI] [PubMed] [Google Scholar]
- 57.Tudor, M., S. Akbarian, R. Z. Chen, and R. Jaenisch. 2002. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes. Proc. Natl. Acad. Sci. USA 99:15536-15541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Varga-Weisz, P., J. Zlatanova, S. Leuba, G. P. Schroth, and K. van Holde. 1994. Binding of histones H1 and H5 and their globular domains to 4-way junction DNA. Proc. Natl. Acad. Sci. USA 91:3525-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Von Hippel, P. J., and O. Berg. 1989. Facilitated target location in biological systems. J. Biol. Chem. 264:675-678. [PubMed] [Google Scholar]
- 60.Wakefield, R. I., B. O. Smith, X. Nan, A. Free, A. Soteriou, D. Uhrin, A. P. Bird, and P. N. Barlow. 1999. The solution structure of the domain from MeCP2 that binds to methylated DNA. J. Mol. Biol. 291:1055-1065. [DOI] [PubMed] [Google Scholar]
- 61.Woodcock, C. L., and R. A. Horowitz. 1998. Electron microscopic imaging of chromatin with nucleosome resolution. Methods Cell Biol. 53:167-186. [DOI] [PubMed] [Google Scholar]
- 62.Yamashita, Y., I. Kondo, T. Fukuda, R. Morishima, A. Kusaga, R. Iwanaga, and T. Matsuishi. 2001. Mutation analysis of the methyl-CpG-binding protein 2 (MECP2) in Rett patients with preserved speech. Brain Dev. 23:(Suppl. 1)S157-S160. [DOI] [PubMed] [Google Scholar]
- 63.Young, J. I., E. P. Hong, J. C. Castle, J. Crespo-Barreto, A. B. Bowman, M. F. Rose, D. Kang, R. Richman, J. M. Johnson, S. Berget, and H. Y. Zoghbi. 2005. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc. Natl. Acad. Sci. USA 102:17551-17558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu, F., J. Thiesen, and W. H. Stratling. 2000. Histone deacetylase-independent transcriptional repression by methyl-CpG-binding protein 2. Nucleic Acids Res. 28:2201-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yusufzai, T. M., and A. P. Wolffe. 2000. Functional consequences of Rett syndrome mutations on human MeCP2. Nucleic Acids Res. 28:4172-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]