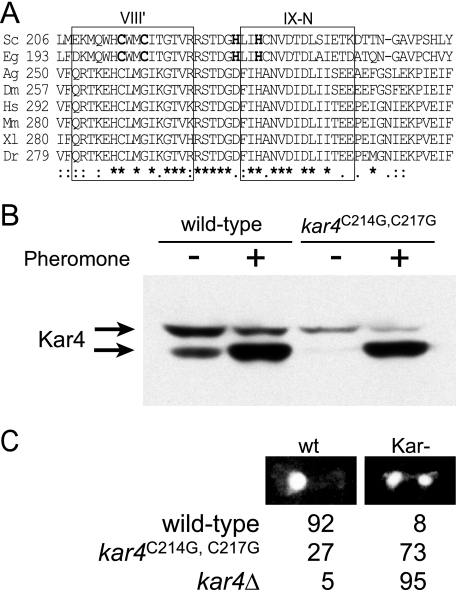
FIG. 2.
A highly conserved region among Kar4 protein family. (A) Pileup sequences of Kar4-related proteins. Family members shown are from Saccharomyces cerevisiae (Sc), Eremothecium gossypii (Eg), Anopheles gambiae (Ag), Drosophila melanogaster (Dm), Homo sapiens (Hs), Mus musculus (Mm), Xenopus laevis (Xl), and Danio rerio (Dr). Identical residues ( ) and similar residues (: or .) are indicated below the pileup. Boxed regions highlight the regions similar to methytransferase domains VIII′ and IX-N (5). Putative zinc-finger ligand residues are indicated in boldface. (B) Mutations in conserved cysteines do not decrease protein levels of Kar4. Western blot showing levels of MS3216 kar4Δ::HIS strain expressing wild-type HA epitope-tagged Kar4 (KAR4::HA on pMR2654) and mutagenized HA epitope-tagged Kar4 (kar4C214G C217G::HA on pMR5511). The cells were grown in the absence (−) or presence (+) of pheromone. The arrows indicate the two forms of Kar4::HAp. (C) Microscopic analysis of mating. Homozygous matings (wild-type × wild-type, kar4C214GC217G × kar4C214GC217G, and kar4Δ × kar4Δ) were mated for 75 min before being fixed in methanol-acetic acid, stained with DAPI to reveal the positions of the nuclei, and examined by fluorescence microscopy. Zygotes were scored as being wild type (nuclei have fused [wt]) or karyogamy defective (nuclei separate and unfused [Kar−]). The percentages of each class are listed.
) and similar residues (: or .) are indicated below the pileup. Boxed regions highlight the regions similar to methytransferase domains VIII′ and IX-N (5). Putative zinc-finger ligand residues are indicated in boldface. (B) Mutations in conserved cysteines do not decrease protein levels of Kar4. Western blot showing levels of MS3216 kar4Δ::HIS strain expressing wild-type HA epitope-tagged Kar4 (KAR4::HA on pMR2654) and mutagenized HA epitope-tagged Kar4 (kar4C214G C217G::HA on pMR5511). The cells were grown in the absence (−) or presence (+) of pheromone. The arrows indicate the two forms of Kar4::HAp. (C) Microscopic analysis of mating. Homozygous matings (wild-type × wild-type, kar4C214GC217G × kar4C214GC217G, and kar4Δ × kar4Δ) were mated for 75 min before being fixed in methanol-acetic acid, stained with DAPI to reveal the positions of the nuclei, and examined by fluorescence microscopy. Zygotes were scored as being wild type (nuclei have fused [wt]) or karyogamy defective (nuclei separate and unfused [Kar−]). The percentages of each class are listed.