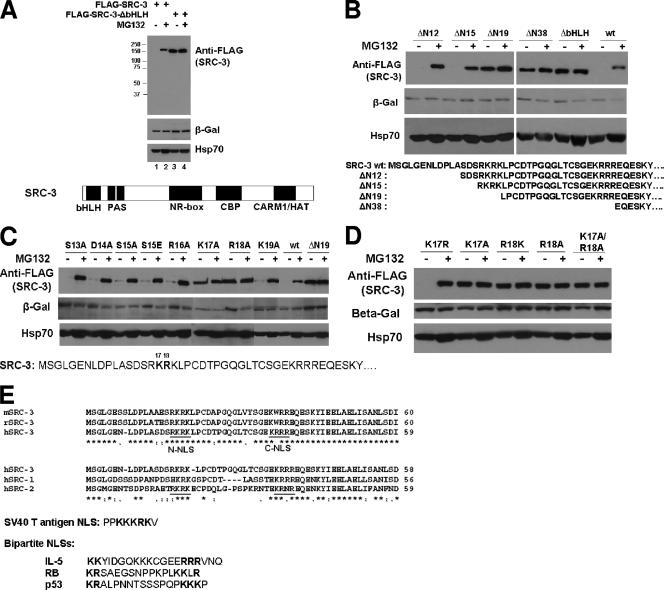
FIG. 1.
Two residues of the SRC-3 bHLH domain essential for SRC-3's proteasome-dependent stability. (A) The bHLH domain of SRC-3 is essential for SRC-3's proteasome-dependent stability. 293 cells were transfected with 200 ng of the plasmids for wt FLAG-SRC-3 (FLAG-SRC-3) or a mutant with deletion of bHLH (FLAG-SRC-3-ΔbHLH) as well as 20 ng of the plasmid for β-Gal. After treatment with MG132 (+) or DMSO (−), cells were harvested for Western analysis using antibodies as indicated. Protein molecular mass markers (in kilodaltons) are shown at the left. The SRC-3 protein structure is indicated at the bottom. (B) Deletion mapping of the bHLH domain. The procedure was the same as that described for panel A, except a series of different SRC-3 deletion mutants was used as indicated. The bottom panel shows the N-terminal sequence of SRC-3 for the deletion mutants. (C) Point mutation analysis of the bHLH domain. The procedure was the same as that described for panel A, except a series of single amino acid point mutants of SRC-3 was used as indicated. The bottom panel shows the sequence position of each point mutant. (D) The procedure was the same as that described for panel A, except additional single point mutants and a double mutant, the K17A/R18A mutant, were used. (E) Sequence analysis. The top panel shows a sequence alignment of the mouse, rat, and human SRC-3 (mSRC-3, rSRC-3, and hSRC-3, respectively) N termini. Underlined putative NLS are labeled N-NLS and C-NLS. The middle panel shows a comparison between hSRC-1, hSRC-2, and hSRC-3, in addition to SV40 T antigen NLS. The bottom panel shows the bipartite NLS of interleukin-5 (IL-5), retinoblastoma (RB), and p53.