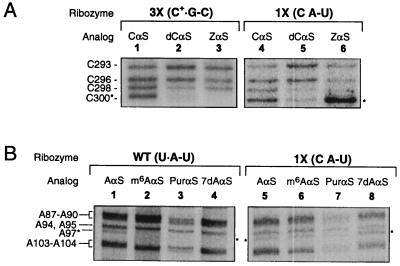
Figure 4.
Nucleotide analog interference mapping experiments. Shown here is a sample set of denaturing polyacrylamide gels that resolve the active ribozyme I2-cleaved RNA fragments resulting from NAIM experiments. Each gel lane is labeled to indicate the specific mutation and the incorporated nucleotide analog in each ribozyme. Nucleotide numbers are indicated to the left of the autoradiographs. Asterisks mark bands of interest. (A) Interference at position 300 in the 3X and 1X mutant ribozymes. Lanes: 1 and 4, CαS phosphorothioate interference controls; 2 and 5, 3X and 1X ribozyme dCαS interference patterns; 3 and 6, 3X and 1X ribozyme ZαS interference patterns. (B) Interference at position 97 in the wild-type and 1X mutant ribozymes. Lanes: 1 and 5, AαS controls; 2, 3, and 4, wild-type m6AαS, PurαS, and 7dAαS interference; 6, 7, and 8, 1X mutant ribozyme m6AαS, PurαS, and 7dAαS interference suppression. The 5′-end labeled control reactions are not shown [similar data have been previously published (11)].