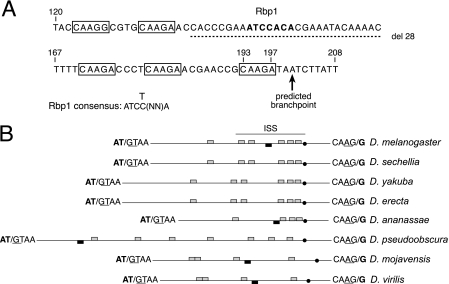
FIG. 2.
Repeated sequences in the ISS are conserved between Drosophila species. (A) The sequence of the ISS element is shown with CAAGR repeats boxed. The predicted branch point for the M1 intron is indicated by an arrow. The 28-nt deletion of nonrepeat sequences is indicated by the dashed line. The numbers above the sequence indicate distance to the 5′ end of the intron. The predicted Rbp1 binding site in the ISS is indicated in boldface type, and the consensus sequence for type B Rbp1 binding sites (12) is shown below. (B) Schematic showing the relative position of CAAG repeats (gray boxes) and potential Rbp1 binding sites (black boxes) in the M1 introns of all Drosophila species examined. All introns are represented to scale (the D. melanogaster M1 intron is 232 nt in length), with the exception of the intron from Drosophila pseudoobscura, which is 758 nt long. Sequences at exon-intron junctions are shown. Splice sites are indicated by a slash mark, and canonical splicing signals are underlined. The translation initiation codon split by M1 is indicated in boldface type. The solid dots indicate the predicted branch point of each intron. The position of the ISS sequences defined by experiments in the D. melanogaster M1 intron is indicated by a line above the diagram.